Abstract
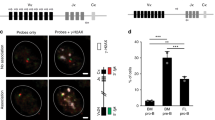
In B lymphopoiesis, activation of the pre-B cell antigen receptor (pre-BCR) is associated with both cell cycle exit and Igk recombination. Yet how the pre-BCR mediates these functions remains unclear. Here, we demonstrate that the pre-BCR initiates a feed-forward amplification loop mediated by the transcription factor interferon regulatory factor 4 and the chemokine receptor C-X-C motif chemokine receptor 4 (CXCR4). CXCR4 ligation by C-X-C motif chemokine ligand 12 activates the mitogen-activated protein kinase extracellular-signal-regulated kinase, which then directs the development of small pre- and immature B cells, including orchestrating cell cycle exit, pre-BCR repression, Igk recombination and BCR expression. In contrast, pre-BCR expression and escape from interleukin-7 have only modest effects on B cell developmental transcriptional and epigenetic programs. These data show a direct and central role for CXCR4 in orchestrating late B cell lymphopoiesis. Furthermore, in the context of previous findings, our data provide a three-receptor system sufficient to recapitulate the essential features of B lymphopoiesis in vitro.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The sequences, ATAC-Seq data for WT small pre-B cells, mb1-Cre+Cxcr4fl/fl (Cxcr4 knockout) small pre-B cells, +IL-7, −IL-7 and −IL-7+CXCL12 cultured WT pre-B cells, and −IL-7 and −IL-7+CXCL12 cultured Cxcr4−/− small pre-B cells, as well as all of the corresponding RNA-Seq data, including for +IL-7+CXCL12 and −IL-7+CXCL12+ERK-i cultured pre-B cells, are deposited in the GenBank database (accession number GSE129311).
References
Clark, M. R., Mandal, M., Ochiai, K. & Singh, H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat. Rev. Immunol. 14, 69–80 (2014).
Herzog, S., Reth, M. & Jumaa, H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol. 9, 195–205 (2009).
Clark, M. R., Cooper, A. B., Wang, L. & Aifantis, I. The pre-B cell receptor in B cell development: recent advances, persistent questions and conserved mechanisms. Curr. Top. Microbiol. Immunol. 290, 87–104 (2005).
Cooper, A. B. et al. A unique function for cyclin D3 in early B cell development. Nat. Immunol. 7, 489–497 (2006).
Lazorchak, A. S. et al. Sin1-mTORC2 suppresses rag and il7r gene expression through Akt2 in B cells. Mol. Cell 39, 433–443 (2010).
Rathmell, J. C. et al. Akt-directed glucose matabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol. Cell Biol. 23, 7315–7328 (2003).
Gottlob, K. et al. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 15, 1406–1418 (2001).
Mandal, M. et al. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat. Immunol. 10, 1110–1117 (2009).
Mandal, M. et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12, 1212–1220 (2011).
Mandal, M. et al. Histone reader BRWD1 targets and restricts recombination to the Igk locus. Nat. Immunol. 16, 1094–1103 (2015).
Herzog, S. et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat. Immunol. 9, 623–631 (2008).
Amin, R. H. & Schlissel, M. S. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 9, 613–622 (2008).
Heizmann, B., Kastner, P. & Chan, S. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J. Exp. Med. 210, 2823–2832 (2013).
Lin, Y. C. et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11, 635–643 (2010).
Sakamoto, S. et al. E2A and CBP/p300 act in synergy to promote chromatin accessibility of the immunoglobulin κ locus. J. Immunol. 188, 5547–5560 (2012).
Van Loo, P. F., Dingjan, G. M., Maas, A. & Hendriks, R. W. Surrogate-light-chain silencing is not critical for the limitation of pre-B cell expansion but is for the termination of constitutive signaling. Immunity 27, 468–480 (2007).
Fistonich, C. et al. Cell circuits between B cell progenitors and the IL-7+ mesenchymal progenitor cells control B cell development. J. Exp. Med. 215, 2596–2599 (2018).
Tokoyoda, K., Egawa, T., Sugiyama, T., Choi, B. I. & Nagasawa, T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity 20, 335–344 (2004).
Johnson, K. et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity 28, 335–345 (2008).
Tian, Y. et al. CXCL12 induces migration of oligodendrocyte precursor cells through the CXCR4-activated MEK/ERK and PI3K/AKT pathways. Mol. Med. Rep. 18, 4374–4380 (2018).
Song, Z. Y., Wang, F., Cui, S. X., Gao, Z. H. & Qu, X. J. CXCR7/CXCR4 heterodimer-induced histone demethylation: a new mechanism of colorectal tumorigenesis. Oncogene 38, 1560–1575 (2019).
Pozzobon, T., Goldoni, G., Viola, A. & Molon, B. CXCR4 signaling in health and disease. Immunol. Lett. 177, 6–15 (2016).
Zheng, N. et al. CXCR7 is not obligatory for CXCL12-CXCR4-induced epithelial-mesenchymal transition in human ovarian cancer. Mol. Carcinog. 58, 144–155 (2019).
Chatterjee, S., Behnam Azad, B. & Nimmagadda, S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 124, 31–82 (2014).
Trampont, P. C. et al. CXCR4 acts as a costimulator during thymic β-selection. Nat. Immunol. 11, 162–170 (2010).
Amin, R. H. et al. Biallelic, ubiquitous transcription from the distal germline Igκ locus promoter during B cell development. Proc. Natl Acad. Sci. USA 106, 522–527 (2009).
Hobeika, E. et al. Testing gene function early in the B cell lineage in mb1-Cre mice. Proc. Natl Acad. Sci. USA 103, 13789–13794 (2006).
Derudder, E. et al. Development of immunoglobulin λ-chain-positive B cells, but not editing of immunoglobulin κ-chain, depends on NF-κB signals. Nat. Immunol. 10, 647–454 (2009).
Ochiai, K. et al. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat. Immunol. 13, 300–307 (2012).
Lagergren, A. et al. The Cxcl12, periostin, and Ccl9 genes are direct targets for early B-cell factor in OP-9 stroma cells. J. Biol. Chem. 282, 14454–14462 (2007).
Mandal, M. et al. BRWD1 orchestrates epigenetic landscape of late B lymphopoiesis. Nat. Commun. 9, 3888–3902 (2018).
Essafi, A. et al. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene 24, 2317–2329 (2005).
Nodland, S. E. et al. IL-7R expression and IL-7 signaling confer a distinct phenotype on developing human B-lineage cells. Blood 118, 2116–2127 (2011).
Malin, S. et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat. Immunol. 11, 171–179 (2010).
Bossen, C. et al. The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat. Immunol. 16, 775–784 (2015).
Montresor, A. et al. CXCR4- and BCR-triggered integrin activation in B-cell chronic lymphocytic leukemia cells depends on JAK2-activated Bruton’s tyrosine kinase. Oncotarget 9, 35123–35140 (2018).
Thompson, E. C. et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity 26, 335–344 (2007).
Vikstrom, I. B. et al. MCL-1 is required throughout B-cell development and its loss sensitizes specific B-cell subsets to inhibition of BCL-2 or BCL-XL. Cell Death Dis. 7, e2345 (2016).
Ma, S. et al. Ikaros and Aiolos inhibit pre-B cell proliferation by directly suppressing c-Myc expression. Mol. Cell Biol. 30, 4149–4158 (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Acknowledgements
We thank M. Olson, R. Ladd and D. Leclerc for cell-sorting services, and the ImmGen Consortium for data assembly. We thank H. Singh (University of Pittsburgh) for providing the PAX5 and FOXO1 ChIP-Seq data, and M. Schlissel (University of Michigan) for providing the Cκ-YFP reporter mice. This work is supported by US National Institutes of Health grants AI120715, AI128785 and AI143778 (to M.R.C.), AI120715-02 and AI128785-01A1 (to M.M.), F32AI143120 (to D.E.K.), T32GM007281 (to K.C.M.), UL1TR002003 (to M.M.-C.) and T32HD007009 (to M.K.O.). Part of the bioinformatics analysis was performed by the UIC Research Informatics Core, supported in part by NCATS through grant UL1TR002003.
Author information
Authors and Affiliations
Contributions
M.M. and M.R.C. designed the experiments. M.M. carried out and analyzed most of the experiments. M.M. and M.M.-C. analyzed the high-throughput sequencing data. M.K.O and D.E.K. assisted with the ATAC-Seq, RNA-Seq and ChIP-Seq experiments, data analyses and flow cytometry. M.V. and K.C.M. performed some sorting and real-time PCR. J.I. and N.K. assisted with the confocal microscopy. I.A. helped to design some of the experiments. F.G. provided the RaDR-GFP mice. M.M. and M.R.C. oversaw the entire project and wrote the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Laurie Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Supplementary Table 1
Rights and permissions
About this article
Cite this article
Mandal, M., Okoreeh, M.K., Kennedy, D.E. et al. CXCR4 signaling directs Igk recombination and the molecular mechanisms of late B lymphopoiesis. Nat Immunol 20, 1393–1403 (2019). https://doi.org/10.1038/s41590-019-0468-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0468-0
This article is cited by
-
BRWD1 orchestrates small pre-B cell chromatin topology by converting static to dynamic cohesin
Nature Immunology (2024)
-
Single-cell technologies in multiple myeloma: new insights into disease pathogenesis and translational implications
Biomarker Research (2023)
-
Disruption to the FOXO-PRDM1 axis resulting from deletions of chromosome 6 in acute lymphoblastic leukaemia
Leukemia (2023)
-
Ebf3+ niche-derived CXCL12 is required for the localization and maintenance of hematopoietic stem cells
Nature Communications (2023)
-
The dynamic functions of IRF4 in B cell malignancies
Clinical and Experimental Medicine (2022)