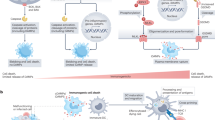
Abstract
The response to systemic infection and injury requires the rapid adaptation of hematopoietic stem cells (HSCs), which proliferate and divert their differentiation toward the myeloid lineage. Significant interest has emerged in understanding the signals that trigger the emergency hematopoietic program. However, the mechanisms that halt this response of HSCs, which is critical to restore homeostasis, remain unknown. Here we reveal that the E3 ubiquitin ligase Speckle-type BTB–POZ protein (SPOP) restrains the inflammatory activation of HSCs. In the absence of Spop, systemic inflammation proceeded in an unresolved manner, and the sustained response in the HSCs resulted in a lethal phenotype reminiscent of hyper-inflammatory syndrome or sepsis. Our proteomic studies decipher that SPOP restricted inflammation by ubiquitinating the innate signal transducer myeloid differentiation primary response protein 88 (MYD88). These findings unearth an HSC-intrinsic post-translational mechanism that is essential for reestablishing homeostasis after emergency hematopoiesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. In addition, raw data generated in this study (bulk RNA-seq, scRNA-seq, ATAC-seq) are available at the GEO database under the accession number GSE112542. Raw data for the proteomics Ms/Ms analysis are available at https://www.ebi.ac.uk/pride/archive/projects/PXD009469.
References
King, K. Y. & Goodell, M. A. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11, 685–692 (2011).
Boettcher, S. & Manz, M. G. Regulation of inflammation- and infection-driven hematopoiesis. Trends Immunol. 38, 345–357 (2017).
Medzhitov, R. Origin and physiological roles of inflammation. Nature 454, 428–435 (2008).
Pietras, E. M. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood 130, 1693–1698 (2017).
Mirantes, C., Passegué, E. & Pietras, E. M. Pro-inflammatory cytokines: emerging players regulating HSC function in normal and diseased hematopoiesis. Exp. Cell Res. 329, 248–254 (2014).
Nagai, Y. et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity 24, 801–812 (2006).
De Luca, K. et al. The TLR1/2 agonist PAM(3)CSK(4) instructs commitment of human hematopoietic stem cells to a myeloid cell fate. Leukemia 23, 2063–2074 (2009).
Essers, M. A. et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 458, 904–908 (2009).
Schuettpelz, L. G. et al. G-CSF regulates hematopoietic stem cell activity, in part, through activation of Toll-like receptor signaling. Leukemia 28, 1851–1860 (2014).
Pietras, E. M. et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18, 607–618 (2016).
Baldridge, M. T., King, K. Y., Boles, N. C., Weksberg, D. C. & Goodell, M. A. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 465, 793–797 (2010).
Ortega-Gomez, A., Perretti, M. & Soehnlein, O. Resolution of inflammation: an integrated view. EMBO Mol. Med. 5, 661–674 (2013).
Serhan, C. N. & Levy, B. D. Resolvins in inflammation: emergence of the pro-resolving superfamily of mediators. J. Clin. Invest. 128, 2657–2669 (2018).
Thompson, B. J. et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 205, 1395–1408 (2008).
King, B. et al. The ubiquitin ligase Huwe1 regulates the maintenance and lymphoid commitment of hematopoietic stem cells. Nat. Immunol. 17, 1312–1321 (2016).
Gao, J. et al. The CUL4-DDB1 ubiquitin ligase complex controls adult and embryonic stem cell differentiation and homeostasis. eLife 4, e07539 (2015).
Strikoudis, A., Guillamot, M. & Aifantis, I. Regulation of stem cell function by protein ubiquitylation. EMBO Rep. 15, 365–382 (2014).
Rape, M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70 (2018).
Bhoj, V. G. & Chen, Z. J. Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437 (2009).
Lara-Astiaso, D. et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014).
Cai, H. & Liu, A. Spop promotes skeletal development and homeostasis by positively regulating Ihh signaling. Proc. Natl Acad. Sci. USA 113, 14751–14756 (2016).
Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. Inducible gene targeting in mice. Science 269, 1427–1429 (1995).
Pietras, E. M. et al. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 211, 245–262 (2014).
Takizawa, H. et al. Pathogen-induced TLR4-TRIF innate immune signaling in hematopoietic stem cells promotes proliferation but reduces competitive fitness. Cell Stem Cell 21, 225–240.e225 (2017).
Delano, M. J. et al. MyD88−dependent expansion of an immature GR-1(+)CD11b(+) population induces T cell suppression and Th2 polarization in sepsis. J. Exp. Med. 204, 1463–1474 (2007).
Passegue, E., Wagner, E. F. & Weissman, I. L. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 119, 431–443 (2004).
Hirai, H. et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat. Immunol. 7, 732–739 (2006).
Olsson, A. et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702 (2016).
Nestorowa, S. et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–e31 (2016).
Giladi, A. et al. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 20, 836–846 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Haas, S. et al. Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell-like megakaryocyte progenitors. Cell Stem Cell 17, 422–434 (2015).
Medzhitov, R. et al. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2, 253–258 (1998).
Tikhonova, A. N. et al. The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019).
Zhuang, M. et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 36, 39–50 (2009).
Cohen, P. TheTLR and IL-1 signalling network at a glance. J. Cell Sci. 127, 2383–2390 (2014).
Akira, S. & Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 (2004).
Gay, N. J., Gangloff, M. & O’Neill, L. A. What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol. 32, 104–109 (2011).
Kaufmann, E. et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell 172, 176–190.e119 (2018).
Netea, M. G. et al. Trained immunity: a program of innate immune memory in health and disease. Science 352, aaf1098 (2016).
Naik, S. et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017).
Li, Y. et al. A functional genomics approach to understand variation in cytokine production in humans. Cell 167, 1099–1110.e1014 (2016).
Ter Horst, R. et al. Host and environmental factors influencing individual human cytokine responses. Cell 167, 1111–1124.e1113 (2016).
Bowman, R. L., Busque, L. & Levine, R. L. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22, 157–170 (2018).
Medyouf, H. The microenvironment in human myeloid malignancies: emerging concepts and therapeutic implications. Blood 129, 1617–1626 (2017).
Barreyro, L., Chlon, T. M. & Starczynowski, D. T. Chronic immune response dysregulation in MDS pathogenesis. Blood 132, 1553–1560 (2018).
Lord, K. A., Hoffman-Liebermann, B. & Liebermann, D. A. Nucleotide sequence and expression of a cDNA encoding MyD88, a novel myeloid differentiation primary response gene induced by IL6. Oncogene 5, 1095–1097 (1990).
Hu, H. & Sun, S. C. Ubiquitin signaling in immune responses. Cell Res. 26, 457–483 (2016).
Heger, K. et al. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature 559, 120–124 (2018).
Wang, Z., Wesche, H., Stevens, T., Walker, N. & Yeh, W. C. IRAK-4 inhibitors for inflammation. Curr. Top. Med. Chem. 9, 724–737 (2009).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Shi, J. et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 (2015).
Pinto do, O. P., Kolterud, A. & Carlsson, L. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 17, 5744–5756 (1998).
Medvar, B., Raghuram, V., Pisitkun, T., Sarkar, A. & Knepper, M. A. Comprehensive database of human E3 ubiquitin ligases: application to aquaporin-2 regulation. Physiol. Genom. 48, 502–512 (2016).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Busino, L. et al. Fbxw7α- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 14, 375–385 (2012).
Florens, L. & Washburn, M. P. Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 328, 159–175 (2006).
Washburn, M. P., Wolters, D. & Yates, J. R. 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 (2001).
McDonald, W. H., Ohi, R., Miyamoto, D. T., Mitchison, T. J. & Yates, J. R. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219, 245–251 (2002).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Tabb, D. L., McDonald, W. H. & Yates, J. R. 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 (2002).
Paoletti, A. C. et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl Acad. Sci. USA 103, 18928–18933 (2006).
Zybailov, B. et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J .Proteome Res. 5, 2339–2347 (2006).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 2921–2929 (2015).
Acknowledgements
We would like to thank all members of the Aifantis laboratory for discussions throughout the duration of this project, specifically E. Wang and B. Aranda for expert advice in CRISPR/Cas9 technology and K. Hockemeyer for expert advice in preparing the manuscript; A. Heguy and the NYU Genome Technology Center (supported in part by National Institutes of Health, National Cancer Institute grant P30CA016087-30) for expertise with sequencing experiments; the NYU Histology Core (5P30CA16087-31) for assistance; C. Loomis and L. Chiriboga for immunohistochemistry experiments; and S. Naik for her intellectual input. This work used computing resources at the High-Performance Computing Facility at the NYU Medical Center. I.A. is supported by the National Institutes of Health (grant nos. R01CA133379, 5R01CA173636, RO1CA216421, RO1CA133379), the Leukemia and Lymphoma Society (TRP Program) and the NYSTEM program of the New York State Health Department (NYSTEM-N11G-255). L.B. is supported in part by grants (nos. R00-CA166181-04 and R01-CA207513-01) from the National Cancer Institute and Gilead Sciences Research Scholars Program in Hematology/Oncology. I.A. dedicates this work to the memory of his mentor H. von Boehmer.
Author information
Authors and Affiliations
Contributions
M.G., I.A. and L.B. conceptualized and designed the study. M.G., I.A. and L.B. prepared the manuscript. M.G. performed, analyzed and interpreted the majority of the experiments describing the mouse modeling. D.O. and L.B. designed, performed and interpreted the majority of the proteomics experiments. I.D., Y.G., L.Z.-R. and A.T. performed all of the computational analysis. N.K. generated mouse strains. Y.D., K.C. and M.M. provided technical assistance with animal models. A.S., L.F. and M.P.W. performed the mass spectrometry. S.T.Y. and K.M.K. performed and interpreted the tissue immunofluorescence and the influenza experiment. C.P. analyzed the mouse pathology. C.B. provided the Spop antibody and shared experimental protocols. A.N.T., K.M.K., C.P. and C.B. provided intellectual input.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Laurie Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Spop is highly expressed in LT-HSC.
a, Top 15 highest expressed ubiquitin ligase genes in LT-HSC. Heatmap represents the normalized counts relative to 10M reads per gene in the indicated populations. The data was collected from Lara-Astiaso., 2016 29 b, Spop expression profile on different hematopoietic cells from the same dataset. c, Immunoblot showing Spop protein levels in the indicated sorted cells. Data are representative from two independent experiments d, Schematic diagram of Spop conditional allele. e, Immunoblot analysis of Spop protein levels of total bone marrow cells from wild-type (Spop+/+MxCre) and KO (SpopΔ/ΔMxCre) mice following a single Poly(I:C) injection. Data are representative of three independent experiments. f, Percentage of mouse weight-change on d15 after Poly (I:C) injection (Spop+/+MxCre: n=9; SpopΔ/ΔMxCre: n=10). Data represent minimum, first quartile, mean, third quartile and maximum. Statistical analysis: unpaired t-test, two-tailed). g, Kaplan-Meier analysis of survival of Spop+/+MxCre and SpopΔ/ΔMxCre hematopoietic chimeras after donor hematopoietic reconstitution and one injection of Poly(I:C) (n=5 wild-type, n=6 Spop KO. Statistical analysis: Mantel-Cox test). h, Hematoxylin & Eosin stained sections of spleen, lung and liver of wild-type (Spop+/+MxCre) and KO (SpopΔ/ΔMxCre) hematopoietic chimeric mice on d21 post Poly (I:C) injection. Scale bars indicate 100 um (Spleen and liver), 1000 um (lung) and 10 um magnifications. Data are representative from 3 independent experiments(pI:C: Poly (I:C). LT-HSC: Lin-c-Kit+Sca-1+Flk2-CD34-. ST-HSC: Lin-c-Kit+Sca-1+Flk2-CD34+. MPP: Lin-c-Kit+Sca-1+Flk2+CD34+. CMP: Lin- c-Kit+ Sca-1+ FcgRIIlowCD34+. GMP: Lin-c-Kit+Sca-1+FcgRIIhighCD34+. MEP: Lin-c-Kit+Sca-1+FcgRII-CD34-. CLPs: Lin-Flk2+Il7R+).
Supplementary Figure 2 Gating strategy.
Representative image of the Flow Cytometry gating strategy to identify B-, T- and myeloid cells in peripheral blood and bone marrow of the mice and HSC and progenitors in the bone marrow of the mice.
Supplementary Figure 3 SpopΔMxCre neutrophils display upregulation of inflammatory response gene programs.
a, GSEA enrichment plots of differentially expressed genes in Spop KO SpopΔ/ΔMxCre neutrophils (n=3 mice) compare to control Spop+/+MxCre (n=2 mice). Statistical significance determined by GSEA Nominal p-value b, Heatmap showing the relative expression of selected Toll-like and NF-kB signaling genes identified to be upregulated in Spop KO compare to controls. c, Heatmap showing the relative expression of selected interferon response genes identified to be upregulated in Spop KO compare to controls. d, Ingenuity upstream analysis of differentially expressed genes in SpopΔMxCre neutrophils compare to control. e, ENRICHR transcription factor analysis of the identified differential upregulated genes in Spop KO neutrophils compare to control. (NES: normalized enrichment score).
Supplementary Figure 4 Granulocytic specific Spop deletion does not promote neutrophilia.
a, Immunoblot analysis of Spop protein levels of bone marrow cKit+ cells of wild type, SpopΔ/ΔMxCre and SpopΔ/ΔCreERT2 mice after Poly(I:C) or Tamoxifen treatment. b, Percentage of myeloid (CD11b+Ly6G+) cells in the peripheral blood of (CD11b+Ly6G+) cells in the peripheral blood of Spop KO and control hematopoietic chimeras on the indicated times after Tamoxifen treatment. Data represent mean±s.d. and dots represent different mice. Statistical analysis: unpaired t-test (two-tailed) c, Representative flow cytometry analysis plots of the proportion of myeloid (CD11b+Ly6G+) cells in the peripheral blood of Spop KO and control hematopoietic chimeras on d15 after a sub-lethal LPS injection. d, Representative flow cytometry analysis plots of the proportion of myeloid (CD11b+Ly6G+) cells in the peripheral blood of Spop KO (and control hematopoietic chimeric mice on the indicated days after intranasal influenza inoculation. e, Spop mRNA relative expression levels in the indicated sorted bone marrow cells. Data represent mean±s.d. (n=3 mice per genotype). The results were first standardized for Gapdh expression levels and then each SpopΔ/Δ sample was expressed as a fraction of the expression detected in the correlated control population from the control littermate. g, Percentage of myeloid (CD11b+Ly6G+) cells in the peripheral blood of the indicated mice on d10 following pIpC-challenge (n=5 per genotype). Data represent mean±s.d. and dots represent different mice. Statistical analysis: unpaired t-test (two-tailed). a-f, Data representative of 3 independent experiments.
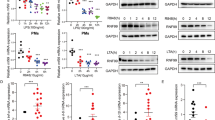
Supplementary Figure 5 Conditional deletion of Spop leads to HSPC expansion and myeloid skewing.
a, Representative flow cytometry analysis plot of Lineage negative bone marrow cells at the indicated days after Poly (I:C) injection. b, Representative flow cytometry analysis plots of the proportion of HSPC populations, including LSK (Lineage-Sca-1+Kit+), LK (Lineage-Sca-1-Kit+), HSC (LSK CD135-, CD48- CD150+), MPP2 (LSK CD135- CD48+ CD150+), MPP3 (LSK CD135- CD48+ CD150-), CMP (LK FcyRlow CD34+), GMP (LK FcyRhigh CD34+), MEP (LK FcyRlo CD34+). c, Percentage of LSK and LK populations in the total bone marrow, percentage of HSC, MPP2, MPP3 and MPP4 in LSKs and percentage of CMP, GMP and MEP in LKs (n=4 mice per genotype. Data represent mean±s.d and dots represent different mice. Statistical analysis: unpaired t-Student, two-tailed). a-d Data are representative from 3 independent experiment. d, Heatmap showing the relative expression of the selected GSEA-analyzed leading edge genes in SpopΔ/ΔMxCre and control sorted LSKs (n=2 mice per genotype) together with control HSC, MPP, CMP, GMP and GN (Granulocyte) wild-type populations from Lara-Astiaso et al., 2016. e, GSEA enrichment plots for HSC, MPP3, MPP4 and granulocytic and macrophage precursor signatures47 in SpopΔ/ΔMxCre and control sorted LSKs (n=2 per genotype). Statistical analysis: Normalized Enrichment Score (NES) and GSEA Nominal p-value. f, Cytokine levels in the serum of the indicated hematopoietic chimeras following one pI:C injection (n=4 per genotype and condition). g, Cytokine levels in the bone marrow fluids of the indicated hematopoietic chimeras following one pI:C injection. f,g Data represent mean±s.d and dots represent different mice. Data are representative of two independent experiments.(h=hours, d=days).
Supplementary Figure 6 Defined gene signature per HSPC clusters.
a,Heatmap showing the expression levels of the 50 most significant markers per cluster, displaying 100 randomly-selected cells of wild-type (Spop+/+MxCre) and Spop KO (SpopΔ/ΔMxCre) LSK. * indicate the upregulation of the C5 specific signature in the cells of other clusters. b, Normalized expression levels of selected population-specific-markers across clusters (C1=4714, C2=2461, C3=1649, C4=1689, C5=1328, C6=1030, C7=196 cells) Data represent mean±s.d. c, Spectral tSNE plot of Spop wild-type (8184) and KO (4920) LSK cells showing the Enrichment Score for Emergency Megakaryocyte gene signature. d, Percentage of Megakarycyte progenitors (Lineage-, Sca-1-, Kit-, CD150+, CD41+) from total progenitor (Lineage-cKit+Sca-1-) cells from the indicated mice (n=4 per genotype and time-point). e, Spectral tSNE plot of Spop wild-type (8184) and KO (4920) LSK cells showing the Enrichment Score for Emergency Granulopoietic gene signature. f, Cell differentiation trajectory using PHATE visualization of wild-type and Spop KO cells, color code for HSC (n=1099), MPP2 (Megakrycyte/Erythroid biased, n=5111), MPP3 (myeloid biased, n=2414) or MPP4 (lymphoid biased, n=3737) gene signatures. g, Spectral tSNE plot of cell cycling color-coded cells. h, Frequency of cells expressing cell cycle genes in SpopΔ/ΔMxCre and control LSKs. i, Percentage of SpopΔ/ΔMxCre and control cell cycling (EdU+) LSKs on d10 post a Poly (I:C) injection. Data represent mean±s.d. n=3. Statistical analysis: unpaired t-student, two-tailed. d,i, Data are representative from 2 independent experiments.
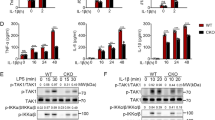
Supplementary Figure 7 SPOP interacts with MYD88 in a phosphorylation independent manner to inhibit IRAK4 signalling.
a, Left, overall view of the SPOP-BTB dimer (pdb access code = 3HTM (Zhuang et al., 2009)) with protomers in green and cyan. Right, overall view of dimer interface rotated 90° in x. The L193 residue on each protomer is shown in red. The structural images were obtained using UCSF chimera software. b, Immunoblot analysis of whole cell lysates from K562 cells stably expressing HA-tagged SPOP(WT) and SPOP(L193P). Cells were treated with cycloheximide (CHX) for the indicated times. EV, empty vector. c, Top, immunoblot analysis of immunoprecipitated WT and truncated forms of FLAG-tagged MYD88 transiently expressed in HEK 293T cells. EV, empty vector. Bottom, diagram showing MYD88 truncations as well as the differential binding to SPOP. d, Immunoblot analysis of immunoprecipitated FLAG-tagged MYD88 and of FLAG-tagged CDC25A, both transiently expressed in HEK293T cells. Equal volumes of the immunopurified protein were both treated with the λ-phosphatase reaction buffer with or without the enzyme. EV, empty vector. e, Immunoblot analysis of immunoprecipitated FLAG-tagged MYD88(WT) and phosphomimetic mutants (S136D, S137D, S136D/S137D) transiently expressed in HEK 293T cells. EV, empty vector. f, In vivo ubiquitylation of immunoprecipitated FLAG-tagged MYD88 upon co-expresison with HA-tagged SPOP and increasing amounts of HA-tagged IRAK4 in HEK293T cells. A low (l.e.) and high exposure (h.e.) are shown. Protein purification was performed in denaturing conditions. g, Immunoblot analysis of immunoprecipitated FLAG-tagged MYD88 co-expressed with HA-tagged SPOP and either HA-tagged IRAK4(WT) or kinase dead (KD) mutant in HEK293T cells. Protein purification was performed in denaturing conditions. h, Immunoblot analysis of whole cell lysates of HPC-7 cells Spop-WT and Spop-KO. Cells were treated with 10 µg/ml lipopolysaccharides (LPS) for the indicated times. A low (l.e.) and high exposure (h.e.) are shown. a-h Data are representative from 3 independent experiments.
Supplementary Figure 8 Spop loss of function reshapes the open chromatin landscape of the HSPCs.
a, Venn-diagram showing the ATAC-seq signals common and unique for wild-type (Spop+/+MxCre n=2 mice) and SpopΔ/ΔMxCre (n=2 mice) LSKs (Lineage-cKit+Sca-1+ bone marrow cells), sorted on d10 following a Poly (I:C) injection (FDR<0.05). b, GREAT Gene Ontology Biological Function analysis of the 25570 SpopΔMxCre differential open chromatin elements. Statistical analysis: GREAT enrichment binomial test from GREAT c, Genome Browser plots showing the normalized ATAC-seq profiles at the promoter and distal elements of the indicated genes for wild-type (blue) and SpopΔMxCre (green) samples. Data are representative from two mice per genotype. d, Ranking of the most enriched transcription factor (TF) motif within SpopΔMxCre vs wild-type differential open chromatin elements (cumulative binomial distribution P<10-10 .Green=TF motif enriched in the Spop(Δ/Δ)MxCre differential open chromatin elements. Blue=TF motif enriched in the Spop+/+MxCre differential open chromatin elements. Black= TF motif enriched in the SpopΔ/ΔMxCre Spop+/+MxCre differential open chromatin elements).Statistical analysis: HOMER enrichment binomial test e, ATAC-seq Footprint visualization for the indicated TF. Aggregated plot of the Tn5 (transposase) insertions counts per nucleotide.
Supplementary Figure 9 Myd88 deficiency restores the steady-state transcriptional program in Spop KO LSKs.
a, Representative flow cytometry analysis plots of the proportion of HSPC in the lineage- bone marrow from hematopoietic chimeric mice following Poly(I:C) injection including: control (Spop+/+MxCre MyD88+/+), Spop KO (SpopΔ/ΔMxCre MyD88+/+), Myd88 KO (Spop+/+MxCre MyD88-/-) and dKO (SpopΔ/ΔMxCre MyD88-/-. Data are representative from 2 independent experiments. b, Spop mRNA expression levels of LSKs on d10 post Poly (I:C) injection (n=2). c, Immunoblot analysis of HSPC cKit+ bone marrow cells of wild-type (Spop+/+MxCre MyD88+/+), Spop KO (SpopΔ/ΔMxCre MyD88+/+) and dKO (Spop+/+MxCre MyD88-/-) mice. Data are representative from 3 independent experiments. d, Heatmap showing the expression levels of the 50 most significant markers per cluster, displaying 100 randomly-selected cells. e, Percentage of Cycling cells per genotype. f, Percentage of myeloid Cd11b+Ly6G+ cells in peripheral blood of SpopΔ/ΔMxCre mice on day 20 following pIpC challenge and antibody treatment. Bar plots represent mean+-s.d, n=4 mice per condition. g, Representative Flow Cytometry Analysis Plots of the proportion of Myeloid (Cd11b+, Ly6G+) cells in the peripheral blood of the pIpC-stimulated mice.
Supplementary information
Supplementary Information
Supplementary Figures 1–9 and unmodified blots
Supplementary Table 1
Ubiquitinome transcriptional profile in HSPCs.
Supplementary Table 2
Differentially expressed genes in Spop KO LSKs compared with controls.
Supplementary Table 3
Population-specific gene expression: mean expression gene per cluster.
Supplementary Table 4
Cluster-specific differentially expressed genes.
Supplementary Table 5
Spop pull-down mass spectrometry.
Supplementary Table 6
Differential open chromatin sites in Spop KO and control LSK 10 days after Poly(I:C).
Supplementary Table 7
Mean gene expression per cluster and genotype.
Rights and permissions
About this article
Cite this article
Guillamot, M., Ouazia, D., Dolgalev, I. et al. The E3 ubiquitin ligase SPOP controls resolution of systemic inflammation by triggering MYD88 degradation. Nat Immunol 20, 1196–1207 (2019). https://doi.org/10.1038/s41590-019-0454-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0454-6
This article is cited by
-
OTUD5 promotes the inflammatory immune response by enhancing MyD88 oligomerization and Myddosome formation
Cell Death & Differentiation (2024)
-
Blockage of CacyBP inhibits macrophage recruitment and improves anti-PD-1 therapy in hepatocellular carcinoma
Journal of Experimental & Clinical Cancer Research (2023)
-
C-type lectin receptor 2d forms homodimers and heterodimers with TLR2 to negatively regulate IRF5-mediated antifungal immunity
Nature Communications (2023)
-
Integrated Analysis of Chromatin and Transcriptomic Profiling Identifies PU.1 as a Core Regulatory Factor in Microglial Activation Induced by Chronic Cerebral Hypoperfusion
Molecular Neurobiology (2023)
-
SPOP negatively regulates Toll-like receptor-induced inflammation by disrupting MyD88 self-association
Cellular & Molecular Immunology (2021)