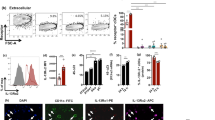
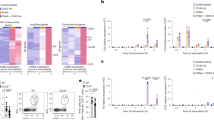
Abstract
Type III interferon (IFN-λ) is important for innate immune protection at mucosal surfaces and has therapeutic benefit against influenza A virus (IAV) infection. However, the mechanisms by which IFN-λ programs adaptive immune protection against IAV are undefined. Here we found that IFN-λ signaling in dendritic cell (DC) populations was critical for the development of protective IAV-specific CD8+ T cell responses. Mice lacking the IFN-λ receptor (Ifnlr1−/−) had blunted CD8+ T cell responses relative to wild type and exhibited reduced survival after heterosubtypic IAV re-challenge. Analysis of DCs revealed IFN-λ signaling directed the migration and function of CD103+ DCs for development of optimal antiviral CD8+ T cell responses, and bioinformatic analyses identified IFN-λ regulation of a DC IL-10 immunoregulatory network. Thus, IFN-λ serves a critical role in bridging innate and adaptive immunity from lung mucosa to lymph nodes to program DCs to direct effective T cell immunity against IAV.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The RNA-seq dataset is available on GEO under accession GSE124399, and the markdown of our analysis of this data is available at https://hemann.galelab.org/.
References
Kotenko, S. V. et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4, 69–77 (2003).
Sheppard, P. et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4, 63–68 (2003).
Prokunina-Olsson, L. et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 45, 164–171 (2013).
Gad, H. H. et al. Interferon-λ is functionally an interferon but structurally related to the interleukin-10 family. J. Biol. Chem. 284, 20869–20875 (2009).
Donnelly, R. P. & Kotenko, S. V. Interferon-lambda: a new addition to an old family. J. Interferon Cytokine Res. 30, 555–564 (2010).
Hemann, E. A., Gale, M. Jr & Savan, R. Interferon lambda genetics and biology in regulation of viral control. Front. Immunol. 8, 1707 (2017).
Lazear, H. M., Nice, T. J. & Diamond, M. S. Interferon-lambda: immune functions at barrier surfaces and beyond. Immunity 43, 15–28 (2015).
Lee, S. & Baldridge, M. T. Interferon-lambda: a potent regulator of intestinal viral infections. Front. Immunol. 8, 749 (2017).
Levy, D. E., Marie, I. J. & Durbin, J. E. Induction and function of type I and III interferon in response to viral infection. Curr. Opin. Virol. 1, 476–486 (2011).
Odendall, C. & Kagan, J. C. The unique regulation and functions of type III interferons in antiviral immunity. Curr. Opin. Virol. 12, 47–52 (2015).
Flannery, B. et al. Interim estimates of 2017–18 seasonal influenza vaccine effectiveness— United States, February 2018. MMWR Morb. Mortal. Wkly. Rep. 67, 180–185 (2018).
Davidson, S. et al. IFNλ is a potent anti-influenza therapeutic without the inflammatory side effects of IFNα treatment. EMBO Mol. Med. 8, 1099–1112 (2016).
Galani, I. E. et al. Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity 46, 875–890.e6 (2017).
Kim, S. et al. The superiority of IFN-λ as a therapeutic candidate to control acute influenza viral lung infection. Am. J. Respir. Cell Mol. Biol. 56, 202–212 (2017).
Klinkhammer, J. et al. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife 7, e33354 (2018).
Broggi, A., Tan, Y., Granucci, F. & Zanoni, I. IFN-λ suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat. Immunol. 18, 1084–1093 (2017).
Crotta, S. et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 9, e1003773 (2013).
Mordstein, M. et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 84, 5670–5677 (2010).
Wang, Y. et al. Involvement of NK cells in IL-28B-mediated immunity against influenza virus infection. J. Immunol. 199, 1012–1020 (2017).
Yin, Z. et al. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J. Immunol. 189, 2735–2745 (2012).
Egli, A. et al. IL-28B is a key regulator of B- and T-cell vaccine responses against influenza. PLoS Pathog. 10, e1004556 (2014).
Koltsida, O. et al. IL-28A (IFN-λ2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol. Med. 3, 348–361 (2011).
Misumi, I. & Whitmire, J. K. IFN-λ exerts opposing effects on T cell responses depending on the chronicity of the virus infection. J. Immunol. 192, 3596–3606 (2014).
Benton, K. A. et al. Heterosubtypic immunity to influenza A virus in mice lacking IgA, all Ig, NKT cells, or gamma delta T cells. J. Immunol. 166, 7437–7445 (2001).
Effros, R. B., Doherty, P. C., Gerhard, W. & Bennink, J. Generation of both cross-reactive and virus-specific T-cell populations after immunization with serologically distinct influenza A viruses. J. Exp. Med. 145, 557–568 (1977).
Sridhar, S. et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 19, 1305–1312 (2013).
Liang, S., Mozdzanowska, K., Palladino, G. & Gerhard, W. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol. 152, 1653–1661 (1994).
Bennink, J., Effros, R. B. & Doherty, P. C. Influenzal pneumonia: early appearance of cross-reactive T cells in lungs of mice primed with heterologous type A viruses. Immunology 35, 503–509 (1978).
Effros, R. B., Bennink, J. & Doherty, P. C. Characteristics of secondary cytotoxic T-cell responses in mice infected with influenza A viruses. Cell. Immunol. 36, 345–353 (1978).
Langlois, R. A., Varble, A., Chua, M. A., Garcia-Sastre, A. & tenOever, B. R. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc. Natl Acad. Sci. USA 109, 12117–12122 (2012).
Waring, B. M. et al. MicroRNA-based attenuation of influenza virus across susceptible hosts. J. Virol. 92, e01741-17 (2018).
Mordstein, M. et al. Interferon-λ contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4, e1000151 (2008).
McGill, J., Van Rooijen, N. & Legge, K. L. Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. J. Exp. Med. 205, 1635–1646 (2008).
Ho, A. W. et al. Lung CD103+ dendritic cells efficiently transport influenza virus to the lymph node and load viral antigen onto MHC class I for presentation to CD8 T cells. J. Immunol. 187, 6011–6021 (2011).
Kim, T. S. & Braciale, T. J. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PloS One 4, e4204 (2009).
Moltedo, B., Li, W., Yount, J. S. & Moran, T. M. Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS Pathog. 7, e1002345 (2011).
Heng, T. S. & Painter, M. W. Immunological genome project C. The immunological genome project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Smith, L. K. et al. Interleukin-10 directly inhibits CD8(+) T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity 48, 299–312.e5 (2018).
Baldridge, M. T. et al. Expression of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of interferon lambda against norovirus and reovirus. J. Virol. 91, e02079-16 (2017).
Abram, C. L., Roberge, G. L., Hu, Y. & Lowell, C. A. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods 408, 89–100 (2014).
Topham, D. J., Tripp, R. A. & Doherty, P. C. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J. Immun. 159, 5197–5200 (1997).
Krishnaswamy, J. K. et al. Migratory CD11b+ conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Sci. Immunol. 2, eaam9169 (2017).
Mount, A. M. et al. Multiple dendritic cell populations activate CD4+ T cells after viral stimulation. PloS One 3, e1691 (2008).
Aparicio-Siegmund, S. & Garbers, C. The biology of interleukin-27 reveals unique pro- and anti-inflammatory functions in immunity. Cytokine Growth Factor Rev. 26, 579–586 (2015).
Llopiz, D. et al. IL-10 expression defines an immunosuppressive dendritic cell population induced by antitumor therapeutic vaccination. Oncotarget 8, 2659–2671 (2017).
Guo, H., Santiago, F., Lambert, K., Takimoto, T. & Topham, D. J. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. J. Virol. 85, 448–455 (2011).
Lazear, H. M. et al. Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci. Transl. Med. 7, 284ra259 (2015).
Ank, N. et al. An important role for type III interferon (IFN-λ/IL-28) in TLR-induced antiviral activity. J. Immunol. 180, 2474–2485 (2008).
Cottey, R., Rowe, C. A. & Bender, B. S. Influenza virus. Curr. Protoc. Immunol. 42, 19.11.1–19.11.32 (2001).
Heaton, N. S., Sachs, D., Chen, C. J., Hai, R. & Palese, P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc. Natl Acad. Sci. USA 110, 20248–20253 (2013).
Schmidt, M. E. et al. Memory CD8 T cells mediate severe immunopathology following respiratory syncytial virus infection. PLoS Pathog. 14, e1006810 (2018).
Moon, J. J. et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007).
Suthar, M. S. et al. The RIG-I-like receptor LGP2 controls CD8+ T cell survival and fitness. Immunity 37, 235–248 (2012).
Daro, E. et al. Polyethylene glycol-modified GM-CSF expands CD11bhighCD11chigh but notCD11blowCD11chigh murine dendritic cells in vivo: a comparative analysis with Flt3 ligand. J. Immunol. 165, 49–58 (2000).
McGill, J. & Legge, K. L. Cutting edge: contribution of lung-resident T cell proliferation to the overall magnitude of the antigen-specific CD8 T cell response in the lungs following murine influenza virus infection. J. Immunol. 183, 4177–4181 (2009).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article 3 (2004).
Loraine, A. E. et al. Analysis and visualization of RNA-Seq expression data using RStudio, bioconductor, and integrated genome browser. Methods Mol. Biol. 1284, 481–501 (2015).
Acknowledgements
We would like to thank the Cell Analysis Facility Flow Cytometry and Imaging Core in the Department of Immunology at the University of Washington and E. Smith for technical assistance. We would like to thank A.M. Kell and M.E. Long for their critical reading of this manuscript. E.A.H. is supported by American Heart Association award 17POST33660907. This work was also supported by National Institutes of Health grants T32AI007509 (E.A.H.), AI132962 (R.A.L.), AI108765 (R.S.), AI104002 (M.G.), AI118916 (M.G.), AI083019 (M.G.) and AI127463 (M.G.).
Author information
Authors and Affiliations
Contributions
E.A.H. designed and performed experiments, analyzed data and wrote the manuscript. R.G. processed and prepared bioinformatic data. J.B.T. performed experiments. R.A.L. provided critical reagents and intellectual input. R.S. provided intellectual input. M.G. designed experiments, provided intellectual input and wrote the manuscript. All authors edited and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 IFN-λ signaling contributes to controlling viral load, but not disease severity, during primary IAV infection. WT and Ifnlr1−/− mice were mock- or IAV-infected with 40 PFU of the mouse-adapted H1N1 strain A/PR/8/34 (PR8)
. a. On days 1, 3, 5, 7, and 9 p.i., lungs were harvested, homogenized, and pulmonary virus titer was determined by plaque assay. Each point represents an individual animal, and data from two, pooled independent experiments are shown. Bars shown mean ± SEM. Day 0 n = 4/group, Day 1 n = 6/group, Day 3 n = 7/group, Day 5 n = 6/group, Day 7 n = 7 (WT) or 6 (Ifnlr1−/−)/group, Day 9 n = 12/group. Significance was determined using one-way ANOVA followed by Tukey’s multiple comparisons test. *indicates p<0.05 b & c. Weight loss (b) and illness score (c) was monitored over 12 days. Dashed line at illness score of 4 indicates presence of respiratory symptoms. n=2 (mock), 7 (WT IAV), or 6 (Ifnlr1−/− IAV) Symbols show mean and error bars represent SD.
Supplementary Figure 2 IFN-λ signaling is not required for development of antibody responses in bronchoalveolar lavage fluid following IAV.
WT and Ifnlr1−/− mice were mock- or IAV-infected with PBS or 40 PFU PR8. Bronchoalveolar lavage fluid was harvested on D35 post infection, and IAV-specific IgM, IgG, IgG1, IgG2a, and IgA antibody levels were determined by ELISA. n=2 (mock) or 4 (IAV) mice/group. Error bars show mean ± SD. Significance was determined using one-way ANOVA followed by Tukey’s multiple comparisons test between WT IAV and Ifnlr1−/− IAV. n.s. indicates p=0.0504, * indicates p<0.05, ** indicates p<0.0001.
Supplementary Figure 3 IFN-λ signaling is dispensable for effector IAV-specific CD4+ T cells responses in the lung.
WT and Ifnlr1−/− mice were mock- or IAV-infected with PBS or 40 PFU PR8 expressing a 2W epitope, respectively. Lungs were harvested on day 9 p.i. and frequency (% of CD4+ T cells) and numbers of NP311- (a) and 2W1S- specific (b) CD4+ T cells were determined by flow cytometry. One representative of two experiments with similar results shown. Bars shown mean ± SD. n=2 (mock) or 3 (IAV) mice/group. c. On day 9 post infection, a portion of the whole lung homogenates were stimulated with media, IAV (NP311) peptide, or PMA + ionomycin for 6 hrs in the presence of brefeldin A. Following stimulation, IFN-γ, TNF, IL-17A, and IL-10 levels were determined by flow cytometry. Two pooled independent experiments are shown with n=6 (WT IAV), or 7 (Ifnlr1−/− IAV) mice/group. Bars show mean ± SEM. Statistical significance determined by one-way ANOVA followed by Tukey’s multiple comparisons test. * indicates p<0.01.
Supplementary Figure 4 BMDCs respond to IFN-λ.
a. WT, Ifnar1−/−, or Ifnlr1−/− BMDCs were left unstimulated or stimulated with 500 ng recombinant murine IFN-λ3 or 10 ng recombinant murine IFNα2 for 1 hr. b. WT BMDCs were stimulated with increasing doses of recombinant murine IFN-λ3 for 30 min. For a and b, following stimulation cells were washed, collected in RIPA buffer, and western blots were performed for phosphorylated STAT1 (Y701), STAT1, and actin. Data shown from one of three representative experiments. c. WT BMDCs were left unstimulated or stimulated with 500 ng recombinant murine IFN-λ3 for 12 hrs. Following stimulation, relative gene expression of Ifi44, Ifit1, Isg15, and Oas3 was determined. Data from three, pooled independent experiments shown where bars represent the mean ± SEM. Statistical significance was determined using unpaired two-sided t-test. * indicates p<0.05, ** indicates p<0.01, *** indicates P<0.001.
Supplementary Figure 5 IFN-λ signaling in LysM+ cells is dispensable for effector IAV-specific CD8+ and CD4+ T cells responses in the lung.
Ifnlr1fl/fl and Ifnlr1fl/flLyz2-cre mice were administered 40 PFU PR8 containing a 2W epitope i.n. a. Weight loss was monitored through day 9 p.i. Symbol shows mean ± SD. b–d. Lungs were harvested on day 9 p.i. and frequency (% of CD8+ or CD4+ T cells) and numbers of PA224-specific CD8+ T cells (b), 2W1S- (c) and NP311-specific (d) CD4+ T cells were assessed by flow cytometry. Data from two pooled experiments shown. For b and c n = 6 mice/group. For d n=5 (Ifnlr1fl/fl) or 6 (Ifnlr1fl/flLyz2-cre) mice/group. For b–d bars show mean ± SEM.
Supplementary Figure 6 IFN-λ signaling in DCs is required for optimal DC function and generation of CD8+ T cell responses capable of mediating protection against heterosubtypic IAV re-challenge.
Following IAV infection, DCs become activated and migrate to initiate IAV-specific CD8+ T cell responses capable of mediating protection against heterosubtypic IAV re-challenge. In Ifnlr1−/−, DC migration and activation are aberrant, and DCs produce IL-10, which correlates with a reduced IAV-specific CD8+ T cell responses that is unable to protect against heterosubtypic IAV re-challenge.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Supplementary Table 1
Rights and permissions
About this article
Cite this article
Hemann, E.A., Green, R., Turnbull, J.B. et al. Interferon-λ modulates dendritic cells to facilitate T cell immunity during infection with influenza A virus. Nat Immunol 20, 1035–1045 (2019). https://doi.org/10.1038/s41590-019-0408-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0408-z
This article is cited by
-
Portraying the dark side of endogenous IFN-λ for promoting cancer progression and immunoevasion in pan-cancer
Journal of Translational Medicine (2023)
-
Interferon antagonists encoded by SARS-CoV-2 at a glance
Medical Microbiology and Immunology (2023)
-
An overview of the crosstalk between YAP and cGAS-STING signaling in non-small cell lung cancer: it takes two to tango
Clinical and Translational Oncology (2022)
-
Interferon lambda in inflammation and autoimmune rheumatic diseases
Nature Reviews Rheumatology (2021)
-
cGAS-STING pathway in cancer biotherapy
Molecular Cancer (2020)