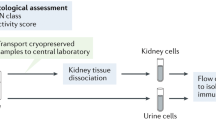
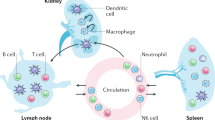
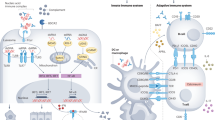
Abstract
Lupus nephritis is a potentially fatal autoimmune disease for which the current treatment is ineffective and often toxic. To develop mechanistic hypotheses of disease, we analyzed kidney samples from patients with lupus nephritis and from healthy control subjects using single-cell RNA sequencing. Our analysis revealed 21 subsets of leukocytes active in disease, including multiple populations of myeloid cells, T cells, natural killer cells and B cells that demonstrated both pro-inflammatory responses and inflammation-resolving responses. We found evidence of local activation of B cells correlated with an age-associated B-cell signature and evidence of progressive stages of monocyte differentiation within the kidney. A clear interferon response was observed in most cells. Two chemokine receptors, CXCR4 and CX3CR1, were broadly expressed, implying a potentially central role in cell trafficking. Gene expression of immune cells in urine and kidney was highly correlated, which would suggest that urine might serve as a surrogate for kidney biopsies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data reported in this publication, including the clinical and serological data of the study participants, are deposited in the ImmPort repository (accession code SDY997). The raw single-cell RNA-seq data are also deposited in dbGAP (accession code phs001457.v1.p1). The processed data can be viewed using an interactive browser at https://immunogenomics.io/ampsle, https://immunogenomics.io/cellbrowser/ and https://portals.broadinstitute.org/single_cell/study/amp-phase-1.
Code availability
All R scripts used to analyze the data reported in this publication are available from the corresponding authors on request.
Change history
13 August 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Costenbader, K. H. et al. Trends in the incidence, demographics, and outcomes of end-stage renal disease due to lupus nephritis in the US from 1995 to 2006. Arthritis Rheum. 63, 1681–1688 (2011).
Narain, S. & Furie, R. Update on clinical trials in systemic lupus erythematosus. Curr. Opin. Rheumatol. 28, 477–487 (2016).
Tektonidou, M. G., Dasgupta, A. & Ward, M. M. Risk of end-stage renal disease in patients with lupus nephritis, 1971–2015: a systematic review and Bayesian meta-analysis. Arthritis Rheumatol. 68, 1432–1441 (2016).
Thanou, A. & Merrill, J. T. Treatment of systemic lupus erythematosus: new therapeutic avenues and blind alleys. Nat. Rev. Rheumatol. 10, 23–34 (2014).
Banchereau, R. et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 165, 1548–1550 (2016).
Liarski, V. M. et al. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci. Transl. Med. 6, 230ra246 (2014).
Hutloff, A. et al. Involvement of inducible costimulator in the exaggerated memory B cell and plasma cell generation in systemic lupus erythematosus. Arthritis Rheum. 50, 3211–3220 (2004).
Kassianos, A. J. et al. Increased tubulointerstitial recruitment of human CD141(hi) CLEC9A(+) and CD1c(+) myeloid dendritic cell subsets in renal fibrosis and chronic kidney disease. Am. J. Physiol. Renal Physiol. 305, F1391–F1401 (2013).
Davidson, A. What is damaging the kidney in lupus nephritis? Nat. Rev. Rheumatol. 12, 143–153 (2016).
Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 (2016).
Hooks, J. J. et al. Immune interferon in the circulation of patients with autoimmune disease. N. Engl. J. Med. 301, 5–8 (1979).
Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Ghebrehiwet, B., Hosszu, K. H. & Peerschke, E. I. C1q as an autocrine and paracrine regulator of cellular functions. Mol. Immunol. 84, 26–33 (2017).
Hulsebus, H. J., O’Conner, S. D., Smith, E. M., Jie, C. & Bohlson, S. S. Complement component C1q programs a pro-efferocytic phenotype while limiting TNFalpha production in primary mouse and human macrophages. Front. Immunol. 7, 230 (2016).
Ikezumi, Y. et al. The sialoadhesin (CD169) expressing a macrophage subset in human proliferative glomerulonephritis. Nephrol. Dial. Transplant. 20, 2704–2713 (2005).
Knutson, M. D. Iron transport proteins: gateways of cellular and systemic iron homeostasis. J. Biol. Chem. 292, 12735–12743 (2017).
Adamson, S. E. et al. Disabled homolog 2 controls macrophage phenotypic polarization and adipose tissue inflammation. J. Clin. Invest. 126, 1311–1322 (2016).
Spadaro, O. et al. IGF1 shapes macrophage activation in response to immunometabolic challenge. Cell Rep. 19, 225–234 (2017).
Varghese, B., Paulos, C. & Low, P. S. Optimization of folate-targeted immunotherapy for the treatment of experimental arthritis. Inflammation 39, 1345–1353 (2016).
Kreslavsky, T. et al. Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and BCR repertoire of B-1a cells. Nat. Immunol. 18, 442–455 (2017).
Angerer, P. et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016).
van den Maaten, L. J. P. H. G. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Dehn, S. & Thorp, E. B. Myeloid receptor CD36 is required for early phagocytosis of myocardial infarcts and induction of Nr4a1-dependent mechanisms of cardiac repair. FASEB J. 32, 254–264 (2017).
Bengsch, B. et al. Deep immune profiling by mass cytometry links human T and NK cell differentiation and cytotoxic molecule expression patterns. J. Immunol. Methods 453, 3–10 (2017).
Bratke, K., Kuepper, M., Bade, B., Virchow, J. C. Jr. & Luttmann, W. Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur. J. Immunol. 35, 2608–2616 (2005).
Boddupalli, C. S. et al. ABC transporters and NR4A1 identify a quiescent subset of tissue-resident memory T cells. J. Clin. Invest. 126, 3905–3916 (2016).
Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016).
Allan, D. S. J. et al. Transcriptome analysis reveals similarities between human blood CD3(−) CD56(bright) cells and mouse CD127(+) innate lymphoid cells. Sci. Rep. 7, 3501 (2017).
McKinney, E. F., Lee, J. C., Jayne, D. R., Lyons, P. A. & Smith, K. G. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616 (2015).
Tilstra, J. S. et al. Kidney-infiltrating T cells in murine lupus nephritis are metabolically and functionally exhausted. J. Clin. Invest. 128, 4884–4897 (2018).
Wherry, E. J. et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007).
Nish, S. A. et al. CD4+ T cell effector commitment coupled to self-renewal by asymmetric cell divisions. J. Exp. Med. 214, 39–47 (2017).
Karnell, J. L. et al. Role of CD11c(+) T-bet(+) B cells in human health and disease. Cell. Immunol. 321, 40–45 (2017).
Jenks, S. A. et al. Distinct effector B cells induced by unregulated Toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity 49, 725–739.e6 (2018).
The FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Lizio, M. et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 16, 22 (2015).
Wang, S. et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat. Commun. 9, 1758 (2018).
Chen, L., Morris, D. L. & Vyse, T. J. Genetic advances in systemic lupus erythematosus: an update. Curr. Opin. Rheumatol. 29, 423–433 (2017).
Chung, S. A. et al. Lupus nephritis susceptibility loci in women with systemic lupus erythematosus. J. Am. Soc. Nephrol. 25, 2859–2870 (2014).
Hua, Z. & Hou, B. TLR signaling in B-cell development and activation. Cell. Mol. Immunol. 10, 103–106 (2013).
Kawasaki, T. & Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 5, 461 (2014).
Madan-Lala, R. et al. Mycobacterium tuberculosis impairs dendritic cell functions through the serine hydrolase Hip1. J. Immunol. 192, 4263–4272 (2014).
Matsuda, S. et al. Regulation of the cell cycle and inflammatory arthritis by the transcription cofactor LBH gene. J. Immunol. 199, 2316–2322 (2017).
Botta, D. et al. Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nat. Immunol. 18, 1249–1260 (2017).
Wing, J. B. et al. A distinct subpopulation of CD25(−) T-follicular regulatory cells localizes in the germinal centers. Proc. Natl Acad. Sci. USA 114, E6400–e6409 (2017).
Winchester, R. et al. Immunologic characteristics of intrarenal T cells: trafficking of expanded CD8+ T cell β-chain clonotypes in progressive lupus nephritis. Arthritis Rheum. 64, 1589–1600 (2012).
Peterson, K. S. et al. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J. Clin. Invest. 113, 1722–1733 (2004).
Davidson, A. Editorial: autoimmunity to vimentin and lupus nephritis. Arthritis Rheumatol. 66, 3251–3254 (2014).
Myles, A., Gearhart, P. J. & Cancro, M. P. Signals that drive T-bet expression in B cells. Cell. Immunol. 321, 3–7 (2017).
Huen, S. C. & Cantley, L. G. Macrophages in renal injury and repair. Ann. Rev. Physiol. 79, 449–469 (2017).
Tabas, I. & Bornfeldt, K. E. Macrophage phenotype and function in different stages of atherosclerosis. Circ. Res. 118, 653–667 (2016).
Auffray, C. et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317, 666–670 (2007).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Nemesh, J. Drop-seq core computational protocol. McCarroll Laboratory http://mccarrolllab.com/wp-content/uploads/2016/03/Drop-seqAlignmentCookbookv1.2Jan2016.pdf (2016).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
McDavid, A. et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461–467 (2013).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300 (1995).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
IUPHAR/BPS. Guide to Pharmacology. http://www.guidetopharmacology.org/download.jsp (2019).
Acknowledgements
This work was supported by the Accelerating Medicines Partnership (AMP) in Rheumatoid Arthritis and Lupus Network. AMP is a public–private partnership (AbbVie Inc., Arthritis Foundation, Bristol-Myers Squibb Company, Foundation for the National Institutes of Health, Janssen Pharmaceuticals, Lupus Foundation of America, Lupus Research Alliance, Merck Sharp & Dohme Corp., National Institute of Allergy and Infectious Diseases, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Pfizer Inc., Rheumatology Research Foundation, Sanofi and Takeda Pharmaceuticals International, Inc.) created to develop new ways of identifying and validating promising biological targets for diagnostics and drug development. Funding was provided through grants from the National Institutes of Health (UH2-AR067676, UH2-AR067677, UH2-AR067679, UH2-AR067681, UH2-AR067685, UH2-AR067688, UH2-AR067689, UH2-AR067690, UH2-AR067691, UH2-AR067694 and UM2-AR067678). N.H. was supported by the David P. Ryan, MD Endowed Chair in Cancer Research. We thank participating Lupus Nephritis Trials Network clinical sites and participants.
Author information
Authors and Affiliations
Consortia
Contributions
A.A., D.A.R, A.D., P.J.H., A.H.J. and D.J.L. analyzed the data. D.A.R., C.C.B., T.M.E., E.P.B., J.A.L. and D.A.H. developed the sample collection and processing protocols. D.A.R., Y.L., P.J.H., A.C., A.N., D.S. and S.S. processed the samples. S.L., D.J.L., A.N., D.S. and S.S. developed the scRNA-seq library preparation protocol. F.Z. and K.S. developed the web-based browser of the data. D.E.S., P.T., E.M., M.D.E., M.P., D.L.K., R.A.F., F.P.S., W.F.P., E.A.M., J.P.B., M.A.P., C.P., K.C.K., E.S.W., D.A.H., D.W. and J.H.A. acquired samples. A.A., D.A.R., C.C.B., A.D., W.A., J.A.L., D.A.H., C.N., D.W., M.K., J.H.A., M.B.B., N.H. and B.D. designed the study. A.D., W.A., D.A.H., C.N., S.R., D.W., M.K., J.H.A., M.B.B., N.H. and B.D. supervised the work. A.A., D.A.R., C.C.B., A.D., P.J.H., N.H. and B.D. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.B.B. is a consultant to Roche in the area of stromal cells.
Integrated supplementary information
Supplementary Figure 1 Analysis of cells obtained from cryopreserved viable kidney tissue.
a, Leukocyte yields from kidney tissue samples processed fresh or after freeze/thaw. b, Leukocyte yields from kidney tissue samples cryopreserved either post-dissociation (frozen cells) or pre-dissociation (frozen tissue). n = 9 segments pooled from 4 different donors. * p < 0.01 by two-tailed Mann-Whitney U-test. c, Cell yields and leukocyte frequencies from kidney samples either cryopreserved or shipped overnight on wet ice. Wilcoxon matched-pairs signed rank test. d, Example of flow cytometry analysis of cells from a cryopreserved lupus nephritis kidney biopsy. e, RNA-seq quality metrics of bulk leukocytes from kidney cells analyzed before (fresh cells) and after freeze/thaw (frozen cells), or obtained from kidney tissue shipped overnight or shipped after tissue cryopreservation (frozen tissue). f-h, The distribution of the number of genes per cell on each processed 384-well plate, in kidney leukocytes (f), kidney epithelial cells (g) and urine leukocytes (h).
Supplementary Figure 2 An overview of the isolated cells.
a-d, Leukocyte yields and composition from LN kidney biopsies, as determined by flow cytometry. a, Number of leukocytes sorted as single cells from LN and control biopsy samples. b, Number of leukocytes sorted from LN biopsies stratified by histologic glomerulonephritis class. c, Flow cytometric assessment of the proportion of leukocytes that are B cells, T cells, monocyte/macrophages, or other in biopsy samples. d, Percentage of B cells among leukocytes in biopsy samples (p = 0.0031; two-tailed Mann-Whitney U-test). e, Projection of the gene expression data of kidney cells following principal component analysis (PCA), in the PC1-PC2 (left) and PC2-PC3 (right) planes. Cells are colored based on their identity as either leukocytes (cyan) or epithelial cells (pink), as determined by flow cytometry. The labeling of clusters in the PC2-PC3 plane is based on the identity of the leading genes in each principal component. f, Same as (e), except cells are colored based on the 384-well plate they were processed on. g, The expression of selected genes in the 10 low-resolution clusters; these were among the genes used to determine the putative lineage of each cell. h, The number of clusters encountered as a function of the number of patients included in the analysis. The presented curve was generated by randomly down-sampling the number of patients analyzed, while counting in each case the number of clusters spanned by the cells of these patients. Patients that had a small number of high quality cells (less than 70) were not included in this analysis, as such patients would lead to a false saturation. Furthermore, to account for the fact that for a cluster to be identified it should contain a sufficient number of cells, we only counted here clusters that had more cells than the smallest cluster identified when analyzing the full cohort (17 cells—cluster CB2b). i, A tSNE plot of the analyzed cells, highlighting the LD controls as opposed to the LN patients. j, A tSNE plot of the analyzed cells, colored by the donor identity.
Supplementary Figure 3 Focused analysis of kidney myeloid cells.
a, The expression of selected genes over the 5 myeloid cell clusters. b-c, The expression of the canonical markers CD16 (FCGR3A) and CD14 on the 5 myeloid clusters. d-f, The changes in the expression of 3 selected genes, along the trajectory from CM0 to CM4; ‘pseudotime’ represents the ordering of the cells along this trajectory. The violin plots (shades) show the distribution of expression levels in equally-spaced intervals along the pseudotime axis (and do not directly correspond to cell clusters). g, The scRNA-seq data of myeloid blood cells sorted from 2 LN patients, projected in the tSNE plane. Low-resolution clustering identified a cluster of CD16+ monocytes; other myeloid cells are grouped here into an ‘other’ set. h, The results of comparing the blood CD16+ monocytes to the 5 myeloid cells clusters found in kidney (CM0-CM4). For each blood cell, the most similar kidney cluster was determined; the bar plot denotes the percentage of myeloid cells most similar to each of the kidney myeloid clusters (only values larger than 0 are shown). i, The increase in the phagocytosis score, computed as the average expression of several genes associated with phagocytosis, in cluster CM1 compared to cluster CM0, in blood myeloid cells. j, Same as (i), but with regard to kidney myeloid cells. ***—p-value < 0.001 (two-tailed Mann-Whitney U-test).
Supplementary Figure 4 Focused analysis of kidney T cells and NK cells.
a, The expression of selected genes across the 7 high-level T/NK cell clusters. b, The expression of selected genes across the two subclusters of cluster CT5, and in comparison to cluster CT1. c, The expression of genes associated with exhaustion, across the different T/NK cell clusters. d, A heatmap showing the separation of LN patients (orange in line above heatmap) and healthy controls (blue), based on expression data of CD8+ T cells sorted from blood samples, and considering a published list of exhaustion markers. Both rows and columns are clustered based on Euclidian distance. Note that out of the 10 LN patients for which blood samples were available, only 8 had sufficient numbers of blood CD8+ T cells to allow sequencing. e, The exhaustion score as calculated in blood CD8+ T cells, comparing the 8 LN patients to the 10 healthy controls. Each point represents a single patient; lines represent the median per group. **—p-value < 0.01 (two-tailed Mann-Whitney U-test). f, The exhaustion score in kidney CD8+ T cells, comparing LN patients to LD controls (same 8 LN patients as in (e)). Each point refers to a cell. g, The expression of selected genes across the two subclusters of cluster CT3. h, The expression of selected genes across the two subclusters of cluster CT0a.
Supplementary Figure 5 Focused analysis of kidney B cells.
a, The expression of selected genes, across the 4 high-level B cell clusters. b, A tSNE projection of all B cells. Note that the cells of cluster CB2 (in cyan) occupy two main separate regions in the tSNE1-tSNE2 plane. c, The expression of selected genes, across the two subclusters of cluster CB2. d-f, The results of classifying the CB2 cells by correlating their gene expression with that of 3 sets of reference samples—FANTOM5 (d), 13 immune cell populations sorted from healthy individuals (Browne et al.; e), and the clusters identified in Villani et al. (f). For each of the 2 subclusters in CB2, the bars denote the percentage of cells most similar to each of the reference samples. For readability, only relevant reference samples are specified, and the rest are collapsed into an ‘other’ category. For all cells in all comparisons, the correlation used for classification was above the corresponding assignability threshold. Note that the reference samples in Villani et al. do not include B cells, and therefore are not relevant for the classification of the cells in cluster CB2a.
Supplementary Figure 6 Focused analysis of cluster C9.
a, The identification of 3 subclusters, the first of which pertains to cells undergoing mitosis. b, The percentage of reads mapped to mitochondrial genes in each subcluster. c, Classification results for each subcluster, comparing the cells of cluster C9 to reference samples found in FANTOM5.
Supplementary Figure 7 Analysis of chemokines, cytokines and their receptors.
a, The distribution of the maximum fraction of expressing cells (taken over all clusters for each receptor), for all receptors. Based on this distribution, a threshold of 0.3 was chosen to define frequently expressed receptors. b–f, The percentage of cells expressing selected chemokine receptors, specified for each cluster. The dashed line in all panels pertains to the above threshold. g–j, The distribution of expression levels of selected chemokines, for each cluster.
Supplementary Figure 8 Analysis of urine cells.
A comparison of gene expression in kidney (x axis) and urine (y axis), for 3 of the clusters represented in both compartments.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8.
Supplementary Table 1: Clinical characteristics of kidney biopsy donors.
Two worksheets are provided—one containing a summary of the clinical characteristics of the studied cohort, the other containing detailed information per sample.
Supplementary Table 2: Sensitivity analysis of clustering parameters.
The results of a detailed sensitivity analysis with regard to the low-resolution clustering step, varying several parameters. Consistency of clustering across various parameter combinations, as defined using the Rand index, is reported.
Supplementary Table 3: Number of cells per cluster and patient.
Rows correspond to patients; columns correspond to clusters.
Supplementary Table 4: Number of cells per cluster and processed 384-well plate.
Rows correspond to plates; columns correspond to clusters.
Supplementary Table 5: Differential expression analysis, comparing LN and LD samples.
Results are reported (in separate worksheets) per cluster, looking only at clusters that had a sufficient number of cells in the LD samples
Supplementary Table 6: Gene lists.
The various gene lists that were used throughout the analysis, each in a separate worksheet.
Supplementary Table 7: Myeloid cells classification by correlation.
The Pearson correlation scores, comparing each kidney myeloid cell (rows) to each reference sample (columns) in Villani et al.
Rights and permissions
About this article
Cite this article
Arazi, A., Rao, D.A., Berthier, C.C. et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol 20, 902–914 (2019). https://doi.org/10.1038/s41590-019-0398-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0398-x
This article is cited by
-
Integrative analysis of single-cell and bulk transcriptome data reveal the significant role of macrophages in lupus nephritis
Arthritis Research & Therapy (2024)
-
The immunoregulatory roles of non-haematopoietic cells in the kidney
Nature Reviews Nephrology (2024)
-
Cytotoxic Tph subset with low B-cell helper functions and its involvement in systemic lupus erythematosus
Communications Biology (2024)
-
Vascular damage in systemic lupus erythematosus
Nature Reviews Nephrology (2024)
-
Joint-specific memory, resident memory T cells and the rolling window of opportunity in arthritis
Nature Reviews Rheumatology (2024)