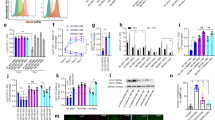
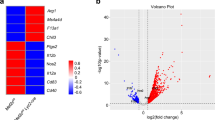
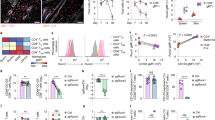
Abstract
Most tissue-resident macrophage populations develop during embryogenesis, self-renew in the steady state and expand during type 2 immunity. Whether shared mechanisms regulate the proliferation of macrophages in homeostasis and disease is unclear. Here we found that the transcription factor Bhlhe40 was required in a cell-intrinsic manner for the self-renewal and maintenance of large peritoneal macrophages (LPMs), but not that of other tissue-resident macrophages. Bhlhe40 was necessary for the proliferation, but not the polarization, of LPMs in response to the cytokine IL-4. During infection with the helminth Heligmosomoides polygyrus bakeri, Bhlhe40 was required for cell cycling of LPMs. Bhlhe40 repressed the expression of genes encoding the transcription factors c-Maf and Mafb and directly promoted expression of transcripts encoding cell cycle-related proteins to enable the proliferation of LPMs. In LPMs, Bhlhe40 bound to genomic sites co-bound by the macrophage lineage-determining factor PU.1 and to unique sites, including Maf and loci encoding cell-cycle-related proteins. Our findings demonstrate a tissue-specific control mechanism that regulates the proliferation of resident macrophages in homeostasis and type 2 immunity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The microarray and ChIP-sequencing data have been deposited in the GEO repository under accession code GSE125730.
References
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
T’Jonck, W., Guilliams, M. & Bonnardel, J. Niche signals and transcription factors involved in tissue-resident macrophage development. Cell. Immunol. 330, 43–53 (2018).
Hashimoto, D. et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Aziz, A., Soucie, E., Sarrazin, S. & Sieweke, M. H. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science 326, 867–871 (2009).
Soucie, E. L. et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science 351, 680–693 (2016).
Imperatore, F. et al. SIRT1 regulates macrophage self-renewal. EMBO J. 36, 2353–2372 (2017).
Rosas, M. et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science 344, 645–648 (2014).
Okabe, Y. & Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832–844 (2014).
Gautier, E. L. et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J. Exp. Med. 211, 1525–1531 (2014).
Jenkins, S. J. et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288 (2011).
Jenkins, S. J. et al. IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. J. Exp. Med. 210, 2477–2491 (2013).
Ruckerl, D. & Allen, J. E. Macrophage proliferation, provenance, and plasticity in macroparasite infection. Immunol. Rev. 262, 113–133 (2014).
Minutti, C. M. et al. Local amplifiers of IL-4Rα-mediated macrophage activation promote repair in lung and liver. Science 356, 1076–1080 (2017).
Bosurgi, L. et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science 356, 1072–1076 (2017).
Gundra, U. M. et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood 123, e110–e122 (2014).
Gundra, U. M. et al. Vitamin A mediates conversion of monocyte-derived macrophages into tissue-resident macrophages during alternative activation. Nat. Immunol. 18, 642–653 (2017).
Kato, Y., Kawamoto, T., Fujimoto, K. & Noshiro, M. DEC1/STRA13/SHARP2 and DEC2/SHARP1 coordinate physiological processes, including circadian rhythms in response to environmental stimuli. Curr. Top. Dev. Biol. 110, 339–372 (2014).
Ow, J. R., Tan, Y. H., Jin, Y., Bahirvani, A. G. & Taneja, R. Stra13 and sharp-1, the non-grouchy regulators of development and disease. Curr. Top. Dev. Biol. 110, 317–338 (2014).
Lin, C. C. et al. IL-1-induced Bhlhe40 identifies pathogenic T helper cells in a model of autoimmune neuroinflammation. J. Exp. Med. 213, 251–271 (2016).
Sun, H. & Taneja, R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc. Natl Acad. Sci. USA 97, 4058–4063 (2000).
St-Pierre, B., Flock, G., Zacksenhaus, E. & Egan, S. E. Stra13 homodimers repress transcription through class B E-box elements. J. Biol. Chem. 277, 46544–46551 (2002).
Li, Y. et al. The expression of antiapoptotic protein survivin is transcriptionally upregulated by DEC1 primarily through multiple sp1 binding sites in the proximal promoter. Oncogene 25, 3296–3306 (2006).
Qian, Y., Zhang, J., Jung, Y. S. & Chen, X. DEC1 coordinates with HDAC8 to differentially regulate TAp73 and DeltaNp73 expression. PLoS One 9, e84015 (2014).
Seimiya, M. et al. Impaired lymphocyte development and function in Clast5/Stra13/DEC1-transgenic mice. Eur. J. Immunol. 34, 1322–1332 (2004).
Kanda, M. et al. Transcriptional regulator Bhlhe40 works as a cofactor of T-bet in the regulation of IFN-γ production in iNKT cells. Proc. Natl Acad. Sci. USA 113, E3394–E3402 (2016).
Kreslavsky, T. et al. Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and BCR repertoire of B-1a cells. Nat. Immunol. 18, 442–455 (2017).
Camponeschi, A. et al. DEC1/STRA13 is a key negative regulator of activation-induced proliferation of human B cells highly expressed in anergic cells. Immunol. Lett. 198, 7–11 (2018).
Martinez-Llordella, M. et al. CD28-inducible transcription factor DEC1 is required for efficient autoreactive CD4+ T cell response. J. Exp. Med. 210, 1603–1619 (2013).
Lin, C. C. et al. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat. Commun. 5, 3551 (2014).
Yu, F. et al. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J. Exp. Med. 215, 1813–1821 (2018).
Huynh, J. P. et al. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J. Exp. Med. 215, 1823–1838 (2018).
Gabryšová, L. & O’Garra, A. Regulating the regulator: Bhlhe40 directly keeps IL-10 in check. J. Exp. Med. 215, 1767–1769 (2018).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Gautier, E. L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Scott, C. L. et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7, 10321 (2016).
Bain, C. C. et al. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat. Commun. 7, 11852 (2016).
Shaw, T. N. et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J. Exp. Med. 215, 1507–1518 (2018).
Kim, K. W. et al. MHC II+ resident peritoneal and pleural macrophages rely on IRF4 for development from circulating monocytes. J. Exp. Med. 213, 1951–1959 (2016).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Gautier, E. L., Ivanov, S., Lesnik, P. & Randolph, G. J. Local apoptosis mediates clearance of macrophages from resolving inflammation in mice. Blood 122, 2714–2722 (2013).
Bertoli, C., Skotheim, J. M. & de Bruin, R. A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell. Biol. 14, 518–528 (2013).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Cain, D. W. et al. Identification of a tissue-specific, C/EBPβ-dependent pathway of differentiation for murine peritoneal macrophages. J. Immunol. 191, 4665–4675 (2013).
Franklin, R. A. & Li, M. O. Ontogeny of tumor-associated macrophages and its implication in cancer regulation. Trends Cancer 2, 20–34 (2016).
Zhu, Y. et al. Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity 47, 323–338 (2017).
Loyher, P. L. et al. Macrophages of distinct origins contribute to tumor development in the lung. J. Exp. Med. 215, 2536–2553 (2018).
Mantovani, A., Marchesi, F., Malesci, A., Laghi, L. & Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017).
Sun, H., Lu, B., Li, R. Q., Flavell, R. A. & Taneja, R. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat. Immunol. 2, 1040–1047 (2001).
Finkelman, F. D. et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J. Immunol. 151, 1235–1244 (1993).
Camberis, M., Le Gros, G. & Urban, J. Jr. Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr. Protoc. Immunol. Chapter 19, Unit 19.12 (2003).
Bando, J. K. et al. The tumor necrosis factor superfamily member RANKL suppresses effector cytokine production in group 3 innate lymphoid cells. Immunity 48, 1208–1219 (2018).
Jablonski, K. A. et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS One 10, e0145342 (2015).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Bailey, T. L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–1659 (2011).
Micallef, L. & Rodgers, P. EulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PLoS One 9, e101717 (2014).
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Disease (NIAID) (AI113118 and AI132653) (B.T.E.) and a Burroughs Wellcome Fund Career Award for Medical Scientists (B.T.E.). N.N.J. was supported by grant 5T32AI007163 from the NIAID. C.-C.L. was supported by the McDonnell International Scholars Academy at Washington University. S.C.-C.H. was supported by the Case Comprehensive Cancer Center American Cancer Society IRG Award (IRG−16–186–21). M.E.C. was supported by the National Science Foundation Graduate Research Fellowship program (DGE-1745038). Research reported in this publication was supported by the Washington University Institute of Clinical and Translational Sciences grant UL1TR002345 from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH. We thank E. Lantelme, A. Cullen and D. Brinja for help with fluorescence-activated cell sorting. We thank W. Beatty and L. Mwaghore for electron microscopy. We thank S. Van Dyken and C. Farnsworth for critical reading of the manuscript. We thank the members of the G. Randolph laboratory for helpful discussions about this project. We thank J. Bando, M. Robinette and T. Ai for help with flow cytometry of the gut.
Author information
Authors and Affiliations
Contributions
N.N.J designed, performed and analyzed experiments and wrote the manuscript. E.A.S., T.R.B., C.-C.L., M.E.C. and C.-W.L. performed experiments. S.C.-C.H. provided reagents, protocols and technical expertise. I.S. and M.N.A. analyzed ChIP-seq data. R.T. provided mice. T.S.S., G.J.R. and J.F.U. provided reagents and technical expertise. B.T.E. designed and analyzed experiments, supervised the studies and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Fig. 1 Bhlhe40 is specifically required in peritoneal and pleural macrophages.
a, Flow cytometry of Bhlhe40GFP transgene reporter expression in pleural macrophages from Bhlhe40GFP+ and Bhlhe40GFP– mice (representative of 2 experiments, n = 3 Bhlhe40GFP+, 2 Bhlhe40GFP-). b-e, Flow cytometry for AMs (b), red pulp macrophages (c), kidney macrophages (d), and Kupffer cells (e) from Bhlhe40+/+ and Bhlhe40-/- mice as in Fig. 1c. f, Flow cytometry for Tim4 expression on LPMs from Bhlhe40+/+ and Bhlhe40-/- mice (representative of 11 experiments, n = 40/group). g, Numbers of Tim4+ LPMs as in f. h, Flow cytometry for CD226 expression on SPMs from Bhlhe40+/+ and Bhlhe40-/- mice (representative of 8 experiments, n = 27/group). i, Numbers of CD226+ SPMs as in h. j, Numbers of peritoneal B cells from Bhlhe40+/+ and Bhlhe40-/- mice (pooled from 12 experiments, n = 35/group). k, Numbers of large pleural macrophages from Bhlhe40+/+ and Bhlhe40-/- mice (pooled from 6 experiments, n = 16 Bhlhe40+/+, 17 Bhlhe40-/-). l, Frequency of Ki67+ large pleural macrophages from Bhlhe40+/+ and Bhlhe40-/- mice (pooled from 3 experiments, n = 7 Bhlhe40+/+, 8 Bhlhe40-/-). m, frequency of 7-AAD+ LPMs from Bhlhe40+/+ and Bhlhe40-/- mice (pooled from 8 experiments, n = 19 Bhlhe40+/+, 18 Bhlhe40-/-). Data in g,i-m are mean ± s.e.m; each symbol represents an individual mouse (g,i-m). ***P < 0.001; ****P < 0.0001, unpaired two-sided Student’s t-test.
Supplementary Fig. 2 Bhlhe40 is cell-intrinsically required in LPMs.
a, Ratio of CD45.1 to CD45.2 cells for small pleural macrophages and pleural B cells from mixed bone marrow chimeras as in Fig. 2d. b, Flow cytometry for the discrimination of donor and recipient LPMs and peritoneal B cells from CD45.1/CD45.2 recipients of transferred peritoneal cells as in Fig. 2k, l. c,d, Gene expression microarray data from Bhlhe40+/+ and Bhlhe40-/- LPMs (in this study) and LysM-Cre- Gata6fl/fl, and LysM-Cre+ Gata6fl/fl LPMs (reanalyzed from 36) were analyzed for shared and unique Bhlhe40 and/or Gata6-dependent genes ( ≥ 2-fold differentially expressed, depicted as a Venn diagram) (c) and differentially expressed genes dependent on both Bhlhe40 and Gata6 (d). e, Gene expression microarray data were analyzed for expression of an LPM gene signature in LPMs and AMs from Bhlhe40+/+ and Bhlhe40-/- mice. Microarray data from LPMs (n = 3/group) and AMs (n = 2/group) are from a single experiment. Data in a are mean ± s.e.m; each symbol represents an individual mouse (a). Significance calculated with an unpaired two-sided Student’s t-test.
Supplementary Fig. 3 Further analysis of responses to IL-4c in Bhlhe40-deficient mice.
a, Flow cytometry of peritoneal macrophage subsets from Bhlhe40+/+, Bhlhe40-/-, and Bhlhe40-/- Il10-/- mice treated with PBS or IL-4c (representative of 2 experiments, n = 3 PBS-treated Bhlhe40+/+, 2 PBS-treated Bhlhe40-/-, 3 PBS-treated Bhlhe40-/- Il10-/-, 5 IL-4c-treated Bhlhe40+/+, 7 IL-4c-treated Bhlhe40-/-, 4 IL-4c-treated Bhlhe40-/- Il10-/-). b, Numbers of LPMs as in a. c, Immunoblotting of cyclins D1-3, cyclin-dependent kinase (CDK) 2, CDK4, CDK6, E2F2, and beta actin in lysates of LPMs from Bhlhe40+/+ and Bhlhe40-/- mice unstimulated or treated with IL-4c (representative of 2 experiments, n = 2/group). d, Frequency of BrdU+ Bhlhe40+/+ (CD45.1), Bhlhe40+/+ (CD45.2), or Bhlhe40-/- (CD45.2) LPMs from mixed bone marrow chimera mice (generated as in Fig. 2a–f) treated with PBS or IL-4c, with LPMs from each donor recovered from the same recipient connected by a line (pooled from 2 experiments, n = 2 PBS-treated [Bhlhe40+/+ (CD45.1) +Bhlhe40+/+ (CD45.2)] and [Bhlhe40+/+ (CD45.1) +Bhlhe40-/- (CD45.2)]; 4 IL-4c-treated [Bhlhe40+/+ (CD45.1) +Bhlhe40+/+ (CD45.2)]; 5 [Bhlhe40+/+ (CD45.1) +Bhlhe40-/- (CD45.2)])). Data in b,d are mean ± s.e.m; (b) each symbol or (d) paired symbols represent an individual mouse. ***P < 0.001, (b) unpaired or (d) paired two-sided Student’s t-test.
Supplementary Fig. 4 Loss of Bhlhe40 causes morphological changes in in vivo IL-4c-stimulated peritoneal macrophages.
a,b, Transmission electron microscopy images of LPMs from naïve (a) and IL-4c-treated (b) Bhlhe40+/+ and Bhlhe40-/- mice (representative of 2 experiments, n = 2 mice (45–50 images)/group). Scale bar, 2 μm. c-h, Transmission electron microscopy for cellular cross-sectional area (c), endoplasmic reticulum (ER) cross-sectional extent (d), ER luminal width (e), vesicle cross-sectional area (f), nucleoli/cross-section (g), mitochondria/cross-section (h) of LPMs from Bhlhe40+/+ and Bhlhe40-/- mice unstimulated or treated with IL-4c (pooled from 2 experiments, n = 2 mice/group [27–60 cells analyzed, except for 17–20 cells analyzed in (d)]). Data in c-h are mean ± s.e.m; each symbol represents an individual cell (c-h). *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, unpaired two-sided Student’s t -test.
Supplementary Fig. 5 Bhlhe40 expression is tightly regulated in resident macrophages.
a, Flow cytometry of Bhlhe40GFP transgene reporter expression in LPMs, SPMs, red pulp macrophages, Kupffer cells, kidney macrophages, and AMs from Bhlhe40GFP+ and Bhlhe40GFP- mice after PBS or IL-4c treatment (representative of 1–2 experiments, n = 3 PBS-treated Bhlhe40GFP+, 1 PBS-treated Bhlhe40GFP-, 5 IL-4c-treated Bhlhe40GFP+, 3 IL-4c-treated Bhlhe40GFP-). b, Numbers of LPMs, red pulp macrophages, Kupffer cells, and AMs from Bhlhe40+/+ and Bhlhe40-/- mice treated with PBS or IL-4c (pooled from 2 experiments, n = 4 PBS-treated Bhlhe40+/+, 3 PBS-treated Bhlhe40-/-, 8 IL-4c-treated Bhlhe40+/+, 7 IL-4c-treated Bhlhe40-/-). c-e, Flow cytometry of BrdU incorporation by red pulp macrophages (c), Kupffer cells (d), and AMs (e) from Bhlhe40+/+ and Bhlhe40-/- mice treated with PBS or IL-4c as in Fig. 5g. Data in b are mean ± s.e.m; each symbol represents an individual mouse (b). *P ≤ 0.05, unpaired two-sided Student’s t-test.
Supplementary Fig. 6 Bhlhe40 is required for normal proliferation of thioglycollate-elicited macrophages during type 2 immunity.
a, Flow cytometry of BrdU incorporation by LPMs and Thio-elicited macrophages from Bhlhe40+/+ and Bhlhe40-/- mice treated with PBS, IL-4c, thioglycollate (Thio), or Thio and IL-4c as in Fig. 5j. b,c, Frequency of pHH3+ LPMs and Thio-elicited macrophages (b) (pooled from 2 experiments, n = 3 PBS-treated Bhlhe40+/+; 4 PBS-treated Bhlhe40-/-; 4 IL-4c-treated and Thio-treated Bhlhe40+/+ and Bhlhe40-/-; 6 Thio and IL-4c-treated Bhlhe40+/+ and Bhlhe40-/-) and RELMα+ LPMs and Thio-elicited macrophages (c) (pooled as in b) from Bhlhe40+/+ and Bhlhe40-/- mice treated with PBS, IL-4c, thioglycollate (Thio), or Thio and IL-4c. Data in b,c are mean ± s.e.m; each symbol represents an individual mouse (b,c). *P ≤ 0.05; ***P < 0.001, unpaired two-sided Student’s t-test.
Supplementary Fig. 7 Bhlhe40 directly regulates gene transcription in LPMs.
a, Gene expression microarray data were analyzed for expression of genes encoding selective regulators of LPM proliferation (Myo18a and C1q) in LPMs from Bhlhe40+/+, Bhlhe40-/-, and LysM-Cre+ Bhlhe40fl/fl mice unstimulated or treated with IL-4c. b-d, Tracings of Bhlhe40 binding, PU.1 binding, and vertebrate conservation at the Il10 (b), Ccl2 (c), and Plac8 (d) loci. e, Bhlhe40-bound, Bhlhe40-dependent genes ( ≥ 2-fold differentially expressed in Bhlhe40+/+ and Bhlhe40-/- LPMs) in LPMs from naïve mice and Bhlhe40-bound, Bhlhe40-dependent genes ( ≥ 2-fold differentially expressed in Bhlhe40+/+ and Bhlhe40-/- LPMs) in LPMs from IL-4c-treated mice, as in Fig. 8i. Underlined genes are highlighted elsewhere in this study. f-h, Tracings of Bhlhe40 binding, PU.1 binding, and vertebrate conservation at the Maf (distal) (f), Mafb (g), and Mafb (distal) (h) loci. LPM Bhlhe40 ChIP-seq data (n = 1/group), microarray data from naïve LPMs (n = 3/group), and microarray data from IL-4c-stimulated LPMs (n = 2/group) are from single separate experiments. LPM PU.1 ChIP-seq data reanalyzed from 35.
Supplementary Fig. 8 Bhlhe40 directly binds to cell cycle-related loci and is required to sustain normal gene expression.
a-d, The proportion of Bhlhe40-bound members of gene sets enriched in Bhlhe40+/+ compared to Bhlhe40-/- LPMs from IL-4c-treated mice. Tracings of Bhlhe40 binding, PU.1 binding, and vertebrate conservation for a representative member of the core enrichment signature for each gene set is presented. Hallmark E2F Targets (a), Hallmark Myc Targets (v1) (b), C5 Chromosome Organization (c), and C5 Cell Cycle Process (d). e-h, GSEA of expression of Bhlhe40-bound genes from gene expression microarray data from LPMs from Bhlhe40+/+ and Bhlhe40-/- mice treated with IL-4c for Hallmark E2F Targets (e), Hallmark Myc Targets v1 (f), C5 Chromosome Organization (g), and C5 Cell Cycle Process (h). C5 Cell Cycle Process is also presented in Fig. 8i. NES, normalized enrichment score. FWER, family-wise error rate. LPM Bhlhe40 ChIP-seq data (n = 1/group) and microarray data (n = 2/group) are from single separate experiments. LPM PU.1 ChIP-seq data reanalyzed from 35. (e-h) NES and FWER.
Supplementary information
Rights and permissions
About this article
Cite this article
Jarjour, N.N., Schwarzkopf, E.A., Bradstreet, T.R. et al. Bhlhe40 mediates tissue-specific control of macrophage proliferation in homeostasis and type 2 immunity. Nat Immunol 20, 687–700 (2019). https://doi.org/10.1038/s41590-019-0382-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0382-5
This article is cited by
-
Bhlhe40 deficiency attenuates LPS-induced acute lung injury through preventing macrophage pyroptosis
Respiratory Research (2024)
-
Genome-wide association and functional interrogation identified a variant at 3p26.1 modulating ovarian cancer survival among Chinese women
Cell Discovery (2021)
-
Thymosin β10 promotes tumor-associated macrophages M2 conversion and proliferation via the PI3K/Akt pathway in lung adenocarcinoma
Respiratory Research (2020)
-
Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment
Nature Communications (2020)
-
Establishment of bone marrow-derived M-CSF receptor-dependent self-renewing macrophages
Cell Death Discovery (2020)