Abstract
Fine control of macrophage activation is needed to prevent inflammatory disease, particularly at barrier sites such as the lungs. However, the dominant mechanisms that regulate the activation of pulmonary macrophages during inflammation are poorly understood. We found that alveolar macrophages (AlvMs) were much less able to respond to the canonical type 2 cytokine IL-4, which underpins allergic disease and parasitic worm infections, than macrophages from lung tissue or the peritoneal cavity. We found that the hyporesponsiveness of AlvMs to IL-4 depended upon the lung environment but was independent of the host microbiota or the lung extracellular matrix components surfactant protein D (SP-D) and mucin 5b (Muc5b). AlvMs showed severely dysregulated metabolism relative to that of cavity macrophages. After removal from the lungs, AlvMs regained responsiveness to IL-4 in a glycolysis-dependent manner. Thus, impaired glycolysis in the pulmonary niche regulates AlvM responsiveness during type 2 inflammation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Code availability
Bioinformatics analyses were performed with publicly available code from bioconductor.org.
Data availability
The data that support the findings of this study are available from the corresponding author upon request. RNA-seq data were deposited at Gene Expression Omnibus, with the following accession code: GSE126309.
References
Hussell, T. & Bell, T. J. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 14, 81–93 (2014).
Van Dyken, S. J. & Locksley, R. M. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu. Rev. Immunol. 31, 317–343 (2013).
Gundra, U. M. et al. Vitamin A mediates conversion of monocyte-derived macrophages into tissue-resident macrophages during alternative activation. Nat. Immunol. 18, 642–653 (2017).
Jenkins, S. J. et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288 (2011).
Jenkins, S. J. et al. IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. J. Exp. Med. 210, 2477–2491 (2013).
Bhatia, S. et al. Rapid host defense against Aspergillus fumigatus involves alveolar macrophages with a predominance of alternatively activated phenotype. PLoS ONE 6, e15943 (2011).
Dai, S., Rajaram, M. V., Curry, H. M., Leander, R. & Schlesinger, L. S. Correction: Fine tuning inflammation at the front door: macrophage complement receptor 3-mediates phagocytosis and immune suppression for Francisella tularensis. PLoS Pathog. 12, e1005504 (2016).
Robbe, P. et al. Distinct macrophage phenotypes in allergic and nonallergic lung inflammation. Am. J. Physiol. Lung Cell Mol. Physiol. 308, L358–L367 (2015).
Reece, J. J., Siracusa, M. C. & Scott, A. L. Innate immune responses to lung-stage helminth infection induce alternatively activated alveolar macrophages. Infect. Immun. 74, 4970–4981 (2006).
Roberts, A. W. et al. Tissue-resident macrophages are locally programmed for silent clearance of apoptotic cells. Immunity 47, 913–927 e916 (2017).
Guilliams, M. et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210, 1977–1992 (2013).
Ginhoux, F. & Guilliams, M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016).
Schneider, C. et al. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 15, 1026–1037 (2014).
Yu, X. et al. The cytokine TGF-beta promotes the development and homeostasis of alveolar macrophages. Immunity 47, 903–912 e904 (2017).
Westphalen, K. et al. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature 506, 503–506 (2014).
Snelgrove, R. J. et al. A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat. Immunol. 9, 1074–1083 (2008).
Gautier, E. L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Gibbings, S. L. et al. Three unique interstitial macrophages in the murine lung at steady state. Am. J. Respir. Cell Mol. Biol. 57, 66–76 (2017).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17, 451–460 (2017).
Bosurgi, L. et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science 356, 1072–1076 (2017).
Minutti, C. M. et al. Local amplifiers of IL-4Ralpha-mediated macrophage activation promote repair in lung and liver. Science 356, 1076–1080 (2017).
Wiesner, D. L., Smith, K. D., Kashem, S. W., Bohjanen, P. R. & Nielsen, K. Different lymphocyte populations direct dichotomous eosinophil or neutrophil responses to pulmonary cryptococcus infection. J. Immunol. 198, 1627–1637 (2017).
Chen, F. et al. An essential role for TH2-type responses in limiting acute tissue damage during experimental helminth infection. Nat. Med. 18, 260–266 (2012).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Gosselin, D. et al. An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222 (2017).
Haczku, A. Protective role of the lung collectins surfactant protein A and surfactant protein D in airway inflammation. J. Allergy Clin. Immunol. 122, 861–879 (2008).
Thawer, S. et al. Surfactant protein-D Is essential for immunity to helminth infection. PLoS Pathog. 12, e1005461 (2016).
Roy, M. G. et al. Muc5b is required for airway defence. Nature 505, 412–416 (2014).
Gollwitzer, E. S. et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 20, 642–647 (2014).
Trompette, A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166 (2014).
Cheung, A. K. et al. Chromosome 14 transfer and functional studies identify a candidate tumor suppressor gene, mirror image polydactyly 1, in nasopharyngeal carcinoma. Proc. Natl Acad. Sci. USA 106, 14478–14483 (2009).
Rodemer, C. et al. Inactivation of ether lipid biosynthesis causes male infertility, defects in eye development and optic nerve hypoplasia in mice. Hum. Mol. Genet. 12, 1881–1895 (2003).
O’Neill, L. A. & Pearce, E. J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016).
Knipper, J. A. et al. Interleukin-4 receptor alpha signaling in myeloid cells controls collagen fibril assembly in skin repair. Immunity 43, 803–816 (2015).
Ip, W. K. E., Hoshi, N., Shouval, D. S., Snapper, S. & Medzhitov, R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017).
Van den Bossche, J., O’Neill, L. A. & Menon, D. Macrophage immunometabolism: where are we (going)? Trends Immunol. 38, 395–406 (2017).
Eming, S. A., Wynn, T. A. & Martin, P. Inflammation and metabolism in tissue repair and regeneration. Science 356, 1026–1030 (2017).
Pelgrom, L. R. & Everts, B. Metabolic control of type 2 immunity. Eur. J. Immunol. 47, 1266–1275 (2017).
Huang, S. C. et al. Metabolic reprogramming mediated by the mTORC2-IRF4 signaling axis Is essential for macrophage alternative activation. Immunity 45, 817–830 (2016).
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Huang, L., Nazarova, E. V., Tan, S., Liu, Y. & Russell, D. G. Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J. Exp. Med. 215, 1135–1152 (2018).
Sinclair, C. et al. mTOR regulates metabolic adaptation of APCs in the lung and controls the outcome of allergic inflammation. Science 357, 1014–1021 (2017).
Gill, S. K. et al. Increased airway glucose increases airway bacterial load in hyperglycaemia. Sci. Rep. 6, 27636 (2016).
Garnett, J. P. et al. Proinflammatory mediators disrupt glucose homeostasis in airway surface liquid. J. Immuno. 189, 373–380 (2012).
Baker, E. H. & BainesD. L. Airway glucose homeostasis: a new target in the prevention and treatment of pulmonary infection. Chest 153, 507–514 (2018).
Mallia, P. et al. Role of airway glucose in bacterial infections in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 142, 815–823 e816 (2018).
Ho, W. E. et al. Metabolomics reveals altered metabolic pathways in experimental asthma. Am. J. Respir. Cell Mol. Biol. 48, 204–211 (2013).
Ostroukhova, M. et al. The role of low-level lactate production in airway inflammation in asthma. Am. J. Physiol. Lung Cell Mol. Physiol. 302, L300–L307 (2012).
Machiels, B. et al. A gammaherpesvirus provides protection against allergic asthma by inducing the replacement of resident alveolar macrophages with regulatory monocytes. Nat. Immunol. 18, 1310–1320 (2017).
Finkelman, F. D. et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine–anti-cytokine antibody complexes. J. Immunol. 151, 1235–1244 (1993).
Cook, P. C. et al. A dominant role for the methyl-CpG-binding protein Mbd2 in controlling Th2 induction by dendritic cells. Nat. Commun. 6, 6920 (2015).
Anderson, K. G. et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9, 209–222 (2014).
Bain, C. C. et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 6, 498–510 (2013).
Phythian-Adams, A. T. et al. CD11c depletion severely disrupts Th2 induction and development in vivo. J. Exp. Med. 207, 2089–2096 (2010).
Wilhelm, C. et al. Critical role of fatty acid metabolism in ILC2-mediated barrier protection during malnutrition and helminth infection. J. Exp. Med. 213, 1409–1418 (2016).
Evans, C. M. et al. The polymeric mucin Muc5ac is required for allergic airway hyperreactivity. Nat. Commun. 6, 6281(2015).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
We thank members of the MCCIR and MacDonald laboratory (University of Manchester) for scientific discussions and some experimental assistance. We thank J. Allen (University of Manchester) for critical reading of the manuscript, the University of Manchester Single Cell Facility for flow cytometry, cell sorting and ImageStream, K. Couper and J. Grainger (University of Manchester) for provision of Pep3 and CX3CR1eGFP mice and M. Travis for providing recombinant TGF-β. This research was supported by a MCCIR PhD studentship (F.R.S.), the Medical Research Council (grant no. MR/P026907/1, H.C. and J.M.), the National Institutes of Health (grant no. HL080396 and HL130938, C.M.E.), the Wellcome Trust Institutional Strategic Support Fund (grant no.105610, R.K.G., D.J.T. and M.Z.K.), Medical Research Foundation UK joint funding with Asthma UK (grant no. MRFAUK-2015–302, T.E.S.), BBSRC studentship (C.S.), a University of Manchester Dean’s Prize Early Career Research Fellowship (P.C.C.), Springboard Award (Academy of Medical Sciences, grant no. SBF002/1076, P.C.C.) and MCCIR core funding (A.S.M. and T.H.). This work was also made possible through use of the Manchester Gnotobiotic Facility that was established with the support of the Wellcome Trust (grant no. 097820/Z/11/B), using founder mice obtained from the Clean Mouse Facility, University of Bern, Switzerland. The Bioimaging Facility microscopes used in this study were purchased with grants from BBSRC, Wellcome Trust and the University of Manchester Strategic Fund.
Author information
Authors and Affiliations
Contributions
P.C.C. and A.S.M. were responsible for conceptualization. F.R.S., S.L.B., M.Z.K., L.C., M.C., G.H., A.C.I. and P.C.C. contributed methodology. F.R.S., S.L.B., M.Z.K., L.C., C.S., M.C., G.H., T.E.S., A.C.I. and P.C.C. conducted investigations. H.C., J.M., C.M.E., T.E.S., D.J.T., R.K.G., D.M.C., T.H. and A.S.M. provided resources. F.R.S., P.C.C. and A.S.M. wrote the first draft. F.R.S., S.L.B., C.M.E., A.C.I., D.J.T., R.K.G., D.M.C., T.H., P.C.C. and A.S.M. reviewed, edited and wrote the final article. D.J.T., R.K.G., D.M.C., T.H., P.C.C. and A.S.M. were involved in funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The Manchester Collaborative Centre for Inflammation Research is a joint venture between the University of Manchester, AstraZeneca and GSK. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Flow cytometry gating strategy for the identification of pulmonary myeloid cells, M(IL-4)-responsive cells and macrophages from various tissue sites.
a, Representative flow cytometry plots showing gating for cells isolated from naïve lung tissue to identify neutrophil (CD11b+Ly6G+ - Ly6G included in the lineage antibody cocktail), IntM (MerTK+CD64+CD11b+Siglec-F-), AlvM (MerTK+CD64+CD11b-Siglec-F+), eosinophil (MerTK-Siglec-F+), dentritic cells (MerTK-CD11c+MHCII+) and monocyte (MerTK-CD64+CD11b+Ly6C+) populations. Data representative of 8 independent experiments. b, RELMα and Ki67 expression, or EdU incorporation, by lung tissue AlvMs and IntMs, or PECMs, on d4 following i.p. PBS or IL-4c administration on d0 and d2, and EdU injection i.p. 3 h before tissue collection. Data representative of 8–10 independent experiments. c, Expression of CD11b and Siglec-F on macrophages (MerTK+CD64+) from PEC, PLEC, liver, colon or lung tissue. Data representative of 2–9 independent experiments.
Supplementary Figure 2 Alveolar macrophages have high basal expression of Ym1, pSTAT6, mTORC1 and mTORC2.
a–d, Histograms of Ym1, pSTAT6, pAkt T308 and pAkt S473 (indicators of mTORC1 and mTORC2 activity, respectively) amounts in lung tissue AlvMs or PECMs from naïve WT or Il4ra-/- mice. Data representative of 2 independent experiments, n = 6 (a), n = 3 (b–d) mice per group. Data analyzed by one-way analysis of variance (ANOVA) with Tukey’s post-test for multiple comparisons, displayed as mean ± s.e.m., *P < 0.05 and **P < 0.01.
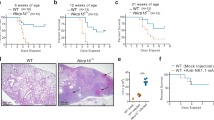
Supplementary Figure 3 Alveolar macrophages are less responsive than are interstitial macrophages during N. brasiliensis infection.
a, Flow cytometry plots showing the gating strategy for cells isolated from lung tissue of naïve mice, or on d2 and d7 following infection s.c. with 500 L3 N. brasiliensis larvae, to identify AlvMs, IntMs and eosinophils. Data representative of 4 independent experiments. b, Flow cytometry plots showing RELMα, Arginase1, Ym1 and Ki67 expression by lung tissue AlvMs and IntMs from naïve or infected mice. Data representative of 1–4 independent experiments. c, Quantification of the percentage of Arginase1+ and Ym1+ lung tissue AlvMs and IntMs from naïve or infected mice, n = 4 (naïve), n = 3 (d2), n = 3 (d7) mice per group. Data analyzed by one-way analysis of variance (ANOVA) with Tukey’s post-test for multiple comparisons, displayed as mean ± s.e.m., *P < 0.05, **P < 0.01 and ****P < 0.0001.
Supplementary Figure 4 Identification of alveolar macrophages based solely on expression of Siglec-F and CD11c may include contaminating interstitial macrophages and eosinophils.
a, Representative flow cytometry plots depicting a traditional gating strategy to identify AlvMs on the basis of Siglec-F and CD11c, showing the proportion of these cells that are AlvMs, IntMs or eosinophils, on the basis of expression of CD64 and MerTK (as defined in the gating strategy in Supplementary Fig. 1a), for cells isolated from lung tissue of naïve mice, or on d2 and d7 following infection s.c. with 500 L3 N. brasiliensis larvae. Data representative of 4 independent experiments. b, Proportion of AlvMs (green) and contaminating IntMs (purple) in the Siglec-F+CD11c+ gate. Data representative of 4 independent experiments, n = 4 (naïve), n = 4 (d2), n = 3 (d7) infected mice per group. c, Proportion of AlvMs, IntMs and eosinophils (red) in the Siglec-F+CD11c+ gate. Data representative of 4 independent experiments, n = 4 (naïve), n = 4 (d2), n = 3 (d7) infected mice per group. d, Representative flow cytometry plots and quantification of CD11c+Siglec-F+ cells that were positive for RELMα. Pie charts demonstrate the significant proportion of these RELMα+ cells that were IntMs or eosinophils, rather than AlvMs. Data pooled from 2 independent experiments, n = 8 (naïve), n = 8 (d2), n = 6 (d7) infected mice per group. e, Representative flow cytometry plots from 4 independent experiments showing FSC and SSC characteristics of gated populations, and overlays demonstrating the inability to remove only eosinophils based solely on high granularity. f, Representative flow cytometry plots from 4 independent experiments depicting overlapping expression of Siglec-F, CD11b and CD11c by AlvMs, IntMs and eosinophils. Data analyzed by one-way analysis of variance (ANOVA) with Tukey’s post-test for multiple comparisons, displayed as mean ± s.e.m., ***P < 0.001.
Supplementary Figure 5 The responses of alveolar and interstitial macrophages to IL-4c are independent of CD200R1, Muc5b and SP-D regulation.
a, CD200R expression on lung tissue AlvMs and IntMs from naïve WT mice. Histograms representative of 3 independent experiments. b, Quantification of the percentage of RELMα+ and Ki67+ lung tissue AlvMs and IntMs from WT or Cd200r1-/- mice on d4 following i.p. PBS or IL-4c administration on d0 and d2. Data representative of 3 independent experiments, n = 4 mice per group (WT AlvM PBS, Cd200r1-/- AlvM PBS, WT AlvM IL-4c, WT IntM PBS, Cd200r1-/- IntM PBS, WT IntM IL-4c), n = 3 mice per group (Cd200r1-/- AlvM IL-4c, Cd200r1-/- IntM IL-4c). c, Representative immunofluorescence images (left) of Muc5b (green) expression in lung sections from WT mice on d4 following i.p. PBS or IL-4c administration on d0 and d2, with quantification of epithelial Muc5b expression (right). Scale bar: 50μM. Data representative of 2 independent experiments, n = 4 (PBS), n = 6 (IL-4c treated) mice per group. d, Quantification of the percentage of RELMα+ and Ki67+ lung tissue AlvMs and IntMs from WT or Muc5b-/- mice on d4 following i.p. PBS or IL-4c administration on d0 and d2. Data representative of 3 independent experiments, n = 4 (WT AlvM PBS, WT IntM PBS), n = 3 (WT AlvM IL-4c, Muc5b-/- AlvM IL-4c, WT IntM IL-4c, Muc5b-/- IntM IL-4c), n = 2 (Muc5b-/- AlvM PBS, Muc5b-/- IntM PBS) mice per group. e, Quantification of the percentage of RELMα+ and Ki67+ lung tissue AlvMs and IntMs from WT or Sfptd-/- mice on d4 following i.p. PBS or IL-4c administration on d0 and d2. Data representative of 3 independent experiments, n = 2 (WT AlvM PBS, WT IntM PBS), n = 3 (WT AlvM IL-4c, WT IntM IL-4c, Sfptd-/- AlvM PBS, Sfptd-/- AlvM IL-4c, Sfptd-/- IntM PBS, Sfptd-/- IntM IL-4c) mice per group. Data analyzed by one-way analysis of variance (ANOVA) with Tukey’s post-test for multiple comparisons, displayed as mean ± s.e.m..
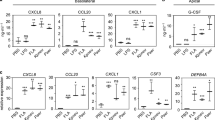
Supplementary Figure 6 Pathway analysis of peritoneal cavity, interstitial and alveolar macrophages from mice given injection of IL-4c versus those given injection of PBS and of alveolar macrophages versus peritoneal cavity macrophages.
KEGG pathway analysis as determined by RNA-seq of significantly altered mRNA transcripts of a, PECMs, b, IntMs or c, AlvMs isolated from PEC or lung tissue by flow cytometry on d4 following i.p. PBS or IL-4c administration on d0 and d2. d, KEGG pathway analysis of significantly altered transcripts of lung tissue AlvMs versus PECMs from IL-4c treated mice, as determined by RNA-seq. Pathways highlighted in bold represent cellular metabolic processes. X-axis: -log10 hypergeometric test (enrichment) P value, line represents the -log10 equivalent of P = 0.05. Data generated from 2–3 independent biological replicates per group, each biological replicate generated from a pool of 3–5 mice.
Supplementary Figure 7 Alveolar macrophages display lower respiratory capacity than that of peritoneal macrophages, and neither fatty acid oxidation nor TGF-β is a vital factor in the recovery of alveolar macrophage M(IL-4) activation.
a, OCR of AlvMs and PECMs isolated from lung tissue or PEC of naïve mice by flow cytometry, at baseline and after sequential treatment (vertical lines) with oligomycin, FCCP, antimycin A and rotenone to measure oxidative phosphorylation (left). Quantification of spare respiratory capacity (right). Graphs show individual replicate wells, n = 3 (AlvM), n = 10 (PECM) wells per group, each group pooled cells from 8 mice. Data representative of 2 independent experiments. b & c, Quantification of Retnla, Arg1 and Chil3 transcript amounts in lung tissue AlvMs from naïve mice cultured for 48 h in media alone or with rIL-4 (20 ngml-1), (b) ± etomoxir (200 µM,) or (c) ± rTGF-β (10 ngml-1) as determined by qPCR. AU arbitrary units. Graphs show individual replicate wells (3 wells per group, each group pooled cells from 8 mice). Data analyzed by one-way analysis of variance (ANOVA) with Tukey’s post-test for multiple comparisons test, displayed as mean ± s.e.m., ***P < 0.001.
Supplementary information
Supplementary Information
Supplementary Figures 1–7 and Supplementary Table 8: Primers for RT-qPCR
Supplementary Table 1: Differentially upregulated expressed genes between PECMs isolated from IL-4c treated mice versus PBS treated mice.
This table displays differentially upregulated expressed genes generated via RNAseq analysis between PECMΦs isolated from IL-4c treated versus PBS-treated mice. The list shows 1,265 significantly upregulated transcripts (P < 0.01).
Supplementary Table 2: Downregulated gene expression between PECMs isolated from IL-4c treated versus PBS treated mice.
This table displays differentially downregulated expressed genes generated via RNAseq analysis between PECMΦs isolated from IL-4c treated versus PBS-treated mice. The list shows 806 significantly downregulated transcripts (P < 0.01).
Supplementary Table 3: Upregulated gene expression between IntMs isolated from IL-4c treated versus PBS treated mice.
This table displays differentially upregulated expressed genes generated via RNAseq analysis between IntMΦs isolated from IL-4c treated versus PBS treated mice. The list shows 80 significantly upregulated transcripts (P < 0.01).
Supplementary Table 4: Downregulated gene expression between IntMs isolated from IL-4c treated versus PBS treated mice.
This table displays differentially downregulated expressed genes generated via RNAseq analysis between IntMΦs isolated from IL-4c treated versus PBS treated mice. The list shows 27 significantly downregulated transcripts (P <0 .01).
Supplementary Table 5. Downregulated gene expression between AlvMs isolated from IL-4c treated versus PBS treated mice.
This table displays differentially down-regulated expressed genes generated via RNAseq analysis between AlvMs isolated from IL-4c treated versus PBS treated mice. The list shows 2 significantly downregulated transcripts (P<0.01).
Supplementary Table 6: Upregulated gene expression between AlvMs versus PECMs isolated from IL-4c treated mice.
This table displays differentially upregulated expressed genes generated via RNAseq analysis between AlvMs versus PECMs isolated from IL-4c treated mice. The list shows the top 1,000 significantly downregulated transcripts (P < 0.01).
Supplementary Table 7: Downregulated gene expression between AlvMs versus PECMs isolated from IL-4c treated mice.
This table displays differentially downregulated expressed genes generated via RNAseq analysis between AlvMs versus PECMs isolated from IL-4c treated mice. The list shows the top 1,000 significantly down-regulated transcripts (P < 0.01).
Rights and permissions
About this article
Cite this article
Svedberg, F.R., Brown, S.L., Krauss, M.Z. et al. The lung environment controls alveolar macrophage metabolism and responsiveness in type 2 inflammation. Nat Immunol 20, 571–580 (2019). https://doi.org/10.1038/s41590-019-0352-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0352-y
This article is cited by
-
MSCs alleviate LPS-induced acute lung injury by inhibiting the proinflammatory function of macrophages in mouse lung organoid–macrophage model
Cellular and Molecular Life Sciences (2024)
-
TREM-1 triggers necroptosis of macrophages through mTOR-dependent mitochondrial fission during acute lung injury
Journal of Translational Medicine (2023)
-
Metabolic heterogeneity of tissue-resident macrophages in homeostasis and during helminth infection
Nature Communications (2023)
-
Harnessing metabolism of hepatic macrophages to aid liver regeneration
Cell Death & Disease (2023)
-
Immune Metabolism in TH2 Responses: New Opportunities to Improve Allergy Treatment — Cell Type-Specific Findings (Part 2)
Current Allergy and Asthma Reports (2023)