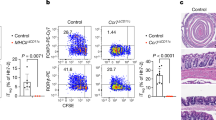
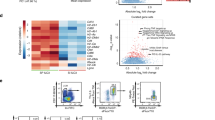
Abstract
Foxp3+ regulatory T cells (Treg cells) are crucial for the maintenance of immune homeostasis both in lymphoid tissues and in non-lymphoid tissues. Here we demonstrate that the ability of intestinal Treg cells to constrain microbiota-dependent interleukin (IL)-17–producing helper T cell (TH17 cell) and immunoglobulin A responses critically required expression of the transcription factor c-Maf. The terminal differentiation and function of several intestinal Treg cell populations, including RORγt+ Treg cells and follicular regulatory T cells, were c-Maf dependent. c-Maf controlled Treg cell–derived IL-10 production and prevented excessive signaling via the kinases PI(3)K (phosphatidylinositol-3-OH kinase) and Akt and the metabolic checkpoint kinase complex mTORC1 (mammalian target of rapamycin) and expression of inflammatory cytokines in intestinal Treg cells. c-Maf deficiency in Treg cells led to profound dysbiosis of the intestinal microbiota, which when transferred to germ-free mice was sufficient to induce exacerbated intestinal TH17 responses, even in a c-Maf-competent environment. Thus, c-Maf acts to preserve the identity and function of intestinal Treg cells, which is essential for the establishment of host–microbe symbiosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Sequencing data supporting the findings of this study have been deposited in the Sequence Read Archive (SRA) database under accession codes SRP123533 (ATAC-Seq) and PRJNA498200 (16S rRNA-Seq) and in the Gene Expression Omnibus (GEO) database under accession code GSE106396 (RNA-Seq).
References
Bunker, J. J. & Bendelac, A. IgA responses to microbiota. Immunity 49, 211–224 (2018).
Dubin, P. J. & Kolls, J. K. Th17 cytokines and mucosal immunity. Immunol. Rev. 226, 160–171 (2008).
Sonnenberg, G. F., Fouser, L. A. & Artis, D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390 (2011).
Ni, J., Wu, G. D., Albenberg, L. & Tomov, V. T. Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 14, 573–584 (2017).
Gratz, I. K. & Campbell, D. J. Organ-specific and memory treg cells: specificity, development, function, and maintenance. Front. Immunol. 5, 333 (2014).
Cretney, E. et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 4, 304–311 (2011).
Vasanthakumar, A. et al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat. Immunol. 16, 276–285 (2015).
Feuerer, M. et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 15, 930–939 (2009).
Arpaia, N. et al. A distinct function of regulatory T cells in tissue protection. Cell 162, 1078–1089 (2015).
Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295 (2013).
Burzyn, D., Benoist, C. & Mathis, D. Regulatory T cells in nonlymphoid tissues. Nat. Immunol. 14, 1007–1013 (2013).
Kühn, R., Löhler, J., Rennick, D., Rajewsky, K. & Muller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993).
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Sellon, R. K. et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 66, 5224–5231 (1998).
Sefik, E. et al. Individual intestinal symbionts induce a distinct population of RORγ+regulatory T cells. Science 349, 993–997 (2015).
Ohnmacht, C. et al. The microbiota regulates type 2 immunity through RORγ+T cells. Science 349, 989–993 (2015).
Kim, K. S. et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 351, 858–863 (2016).
Campbell, C. et al. Extrathymically generated regulatory T cells establish a niche for intestinal border-dwelling bacteria and affect physiologic metabolite balance. Immunity 48, 1245–1257.e9 (2018).
Kawamoto, S. et al. Foxp3+ T cells regulate immunoglobulin A selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41, 152–165 (2014).
Xu, J. et al. c-Maf regulates IL-10 expression during Th17 polarization. J. Immunol. 182, 6226–6236 (2009).
Neumann, C. et al. Role of Blimp-1 in programing Th effector cells into IL-10 producers. J. Exp. Med. 211, 1807–1819 (2014).
Saraiva, M. et al. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity 31, 209–219 (2009).
Apetoh, L. et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 11, 854–861 (2010).
Gabryšová, L. et al. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4+T cells. Nat. Immunol. 19, 497–507 (2018).
Rutz, S. et al. Transcription factor c-Maf mediates the TGF-β-dependent suppression of IL-22 production in T(H)17 cells. Nat. Immunol. 12, 1238–1245 (2011).
Xu, M. et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018).
Wheaton, J. D., Yeh, C.-H. & Ciofani, M. Cutting edge: c-Maf is required for regulatory T cells to adopt RORγt+ and follicular phenotypes. J. Immunol. 199, 3931–3936 (2017).
Pot, C. et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol. 183, 797–801 (2009).
Steward-Tharp, S. M. et al. A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood 123, 2978–2987 (2014).
Madan, R. et al. Nonredundant roles for B cell-derived IL-10 in immune counter-regulation. J. Immunol. 183, 2312–2320 (2009).
Wende, H. et al. The transcription factor c-Maf controls touch receptor development and function. Science 335, 1373–1376 (2012).
Linterman, M. A. et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 17, 975–982 (2011).
Okkenhaug, K. et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3–kinase mutant mice. Science 297, 1031–1034 (2002).
Ciofani, M. et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012).
Chassaing, B., Aitken, J. D., Malleshappa, M. & Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 104, Unit 15.25.–15.25.14 (2014).
Ogawa, A., Andoh, A., Araki, Y., Bamba, T. & Fujiyama, Y. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin. Immunol. 110, 55–62 (2004).
O’Connor, W. et al. A protective function for interleukin 17A in T cell mediated intestinal inflammation. Nat. Immunol. 10, 603–609 (2009).
Sugimoto, K. et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 118, 534–544 (2008).
Zindl, C. L. et al. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc. Natl Acad. Sci. USA 110, 12768–12773 (2013).
Roy, U. et al. Distinct microbial communities trigger colitis development upon intestinal barrier damage via innate or adaptive immune cells. Cell Rep. 21, 994–1008 (2017).
Siewe, L. et al. Interleukin-10 derived from macrophages and/or neutrophils regulates the inflammatory response to LPS but not the response to CpG DNA. Eur. J. Immunol. 36, 3248–3255 (2006).
Amsen, D. et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity 27, 89–99 (2007).
Cossarizza, A. et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur. J. Immunol. 47, 1584–1797 (2017).
Zimmermann, J. et al. High-resolution microbiota flow cytometry reveals dynamic colitis-associated changes in fecal bacterial composition. Eur. J. Immunol. 46, 1300–1303 (2016).
Kau, A. L. et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl. Med. 7, 276ra24–276ra24 (2015).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108, 4516–4522 (2011).
Gálvez, E. J. C., Iljazovic, A., Gronow, A., Flavell, R. & Strowig, T. Shaping of intestinal microbiota in Nlrp6- and Rag2-deficient mice depends on community structure. Cell Rep. 21, 3914–3926 (2017).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217 (2013).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Erben, U. et al. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 7, 4557–4576 (2014).
Wu, T. D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
This research was supported by the Deutsche Forschungsgemeinschaft (DFG) under Germany’s Excellence Strategy—EXC 2167-390884018 (to A.S.), SFB650 (to A.S.) and CRC/TR 241 (to A.S. and C.R.), the German Federal Ministry of Education and Science (BMBF)—Project InfectControl 2020 Projekt DIAT FKZ: 03ZZ0827A (to A.S.), the state of Berlin and the ‘European Regional Development Fund’ ERDF 2014-2020, EFRE 1.8/11, Deutsches Rheuma-Forschungszentrum (to F.H., G.A.H, P.M. and M.F.M), the Helmholtz Association project grant VH-NG-933 (to T. Strowig), the National Health and Medical Research Council (NHMRC) project grants (1069075, 1106378 to A .Kallies), the Sylvia and Charles Viertel Foundation (fellowship to A. Kallies), a Walter and Eliza Institute Centenary Fellowship funded by the CSL (to W.S.), a fellowship by the Deutsche Forschungsgemeinschaft (to J.B.) and the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC Independent Research Institute Infrastructure Support scheme. We thank the DRFZ Flow Cytometry Core Facility and the BCRT Flow Cytometry Lab for cell sorting. In addition, we thank Renee Gloury and Victoria von Götze for technical and experimental help.
Author information
Authors and Affiliations
Contributions
C.N. and J.B. designed and performed most experiments, analyzed the data, generated the figures and wrote the manuscript. U.R. and T. Strowig performed and supervised microbiota studies. P.P.T., Y.L., C.S., P.M. and T. Sidwell performed experiments. A.V. provided discussion and proofread the manuscript. A.B. and H.C. performed and supervised IgA studies. F.H., T.L.A., J.A.H., E.J.C.G., Y.H. and W.S. performed computational analyses of 16 S rRNA-seq, RNA-seq, ChIP-seq and ATAC-seq data. C.E. performed studies with V463 mut-STAT3 mice. G.A.H. and M.F.M. supervised sequencing experiments and provided reagents and equipment for their execution. D.A. and C.R. provided expertise and Notch1floxNotch2floxCd4Cre mice and Il10GFP reporter mice, respectively. A. Kruglov contributed to data interpretation and discussions and assisted in DSS colitis experiments. S.R., A. Kallies and A.S. conceived the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
T.L.A., J.A.H. and S.R. are full-time employees of Genentech and shareholders of Roche.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Gating strategy.
Flow cytometric profiles illustrating the gating strategy for phenotypic analysis of (a) splenic or (b) colonic LP CD4+ TCRβ+ T cells. The presented profiles are representative of at least five independent stainings.
Supplementary Figure 2 Expression of eTreg cell and/or activation markers in c-Maf+ Treg cells versus that of c-Maf− Treg cells.
(a) Flow cytometric analysis shows expression of ICOS, CTLA4, TIGIT, CD69, Blimp-1 and CD25 in splenic c-Maf+ and c-Maf− Foxp3+ Treg cells. Histograms are representative of three independent experiments. (b–c) Flow cytometric profiles of c-Maf versus (b) ICOS and (c) TIGIT expression by Foxp3+ Treg cells from different organs. Contour plots are representative of at least three independent experiments. (d) Expression of c-Maf by MACS-sorted CD4+ CD25+ splenic Treg cells after in vitro stimulation for 72 hours with anti-CD3/CD28 and rIL-2 in the presence or absence of the Notch ligand Dll-4. Histograms (left) show c-Maf expression (gated on Foxp3+ cells). Graph on the right shows frequency of c-Maf+ cells among Foxp3+ Treg cells (pooled data from three independent experiments, mean ± SEM, **P<0.0015). (e) Expression of c-Maf by mLNs Foxp3+ Treg cells from Notch1/2fl/flCd4Cre (N1/2∆CD4) and wild-type SPF (Ctrl) mice. Histograms (left) show c-Maf expression. Graph on the right shows quantification of c-Maf gMFI (n = 4, mean ± SEM). All statistical differences were tested using an unpaired Student’s t-test (two-tailed).
Supplementary Figure 3 Phenotypic characterization of Maf∆Treg mice.
(a) Expression of Il10:GFP by Foxp3+ and Foxp3− CD4+ TCRβ+ T cells isolated from indicated organs of naïve Il10GFP reporter mice as measured by flow cytometry. Representative flow cytometric profiles of Foxp3 versus Il10:GFP expression by splenic and colonic CD4+ TCRβ+ T cells are shown on top. The graph below shows quantification (n = 4, mean ± SEM, *P<0.05, ***P<0.001). (b) Flow cytometric profiles of c-Maf versus Foxp3 expression by CD4+ TCR+ T cells from different organs of Foxp3Cre (Ctrl) and Maffl/flFoxp3Cre (Maf∆Treg) mice. The presented profiles are representative of at least three independent stainings. (c) Frequency of Foxp3+ Treg cells among CD4+ TCRβ+ T cells from indicated organs of Maf∆Treg and control mice (n = 5, mean ± SEM). (d) Enumeration of total cells isolated from different organs of Maf∆Treg and control mice (n = 5, mean ± SEM, *P<0.05, **P<0.01). (e) Flow cytometric analysis shows expression of ICOS, TIGIT, CD69, CD25 in splenic CD62L− Foxp3+ Treg cells of Maf∆Treg and control mice. Histograms are representative of three independent experiments. (f) Flow cytometric analysis of Blimp-1 expression in colonic Foxp3+ Treg cells from Maf∆Treg and control mice. On top, representative contour plots displaying Blimp-1 versus c-Maf expression are shown. Blimp-1-deficient Foxp3+ Treg cells from Prdm1fl/flCd4Cre (Prdm1∆CD4) mice served as a negative control for the Blimp-1 staining. Below, Blimp-1 expression is shown by representative histograms (left) and Blimp-1 gMFI is quantified (right, n = 3, mean ± SEM). (g) Flow cytometric analysis of IRF4 expression in colonic Foxp3+ Treg cells from Maf∆Treg and control mice. On the left, a representative contour plot displaying IRF4 versus c-Maf expression is shown. On the right, IRF4 expression in colonic Foxp3+ Treg cells is shown by representative histograms and IRF4 gMFI is quantified (n = 3, mean ± SEM). Foxp3− conventional T cells (Tcon) from control mice served as a negative control for the IRF4 staining. (h) Flow cytometric analysis of AhR expression in colonic Foxp3+ Treg cells from Maf∆Treg and control mice. On the left, representative contour plots displaying AhR versus c-Maf expression are shown. AhR-deficient Foxp3+ Treg cells from Ahr-/- mice served as a negative control for the AhR staining. On the right, AhR expression is shown by representative histograms and AhR gMFI is quantified (n = 3, mean ± SEM, **P = 0.0097). All statistical differences were tested using an unpaired Student’s t-test (two-tailed).
Supplementary Figure 4 Rorc∆Treg mice have normal intestinal TH17 cell responses.
(a) Flow cytometric analysis of c-Maf expression in different Treg cell subsets from PPs. Histograms are representative of three independent experiments. (b) Frequency of TFR cells (PD1+CXCR5+ cells among Foxp3+ Treg cells) in PPs of Maf∆Treg and control mice. Representative contour plots of PD1 versus CXCR5 expression by Foxp3+ Treg cells are shown on the left. The graph on the right shows quantification (n = 4, mean ± SEM, ***P < 0.0001). (c) Flow cytometric profiles of RORγt versus Foxp3:YFP expression by colonic Foxp3+ Treg cells of mosaic Foxp3Cre/+ or Maffl/flFoxp3Cre/+ female mice. The presented profiles are representative of at least two independent stainings. (d) RORγt (n = 4, mean ± SEM, *P = 0.0462) versus T-bet (n = 5, mean ± SEM, *P = 0.0212) expression by colonic Foxp3− conventional T cells of Maf∆Treg and control mice as measured by flow cytometry. Representative flow cytometric profiles are shown on top. Graphs below show quantification. (e) Contour plots show RORγt and c-Maf expression of Foxp3+ and Foxp3− CD4+TCRβ+ T cells from mLNs of Maf∆Treg and control mice as measured by flow cytometry. The presented plots are representative of at least three independent stainings. (f) Frequencies of IL-17A+ (*P = 0.0092) and IFN-γ+ cells among colonic Foxp3+ Treg cells of Foxp3Cre (Ctrl, n = 4, mean ± SEM) and Rorcfl/flFoxp3Cre (Rorc∆Treg, n = 3, mean ± SEM) mice after ex vivo PMA/ionomycin restimulation as measured by flow cytometry. (g) Frequency of IL-17A+ cells among colonic Foxp3− conventional T cells of Rorc∆Treg (n = 3, mean ± SEM) and control (n = 4, mean ± SEM) mice after ex vivo PMA/ionomycin restimulation as measured by flow cytometry. All statistical differences were tested using an unpaired Student’s t-test (two-tailed).
Supplementary Figure 5 Tissue-specific gene regulation by c-Maf in gut-associated Treg cells.
(a) Foxp3+ Treg cells were sorted from spleen of Maf∆Treg and control mice and subjected to RNA sequencing. MA plot showing comparison of gene expression between c-Maf-deficient and control Treg cells. Genes up-regulated in c-Maf-deficient Treg cells are highlighted in red; genes down-regulated are highlighted in blue (FDR < 0.1). DE analysis was performed using limma. P values were adjusted to control the global false discovery rate (FDR) across all comparisons with the ‘global’ option of the limma package. Data represent the combined analysis of three biologically independent samples. (b) Venn diagram displaying the overlap between genes differentially expressed (DE) between c-Maf-deficient and control Treg cells from LP, PPs and spleens. (c) Heatmap showing relative expression (z-score) of genes that were differentially expressed (FDR < 0.1) between c-Maf-deficient and control Treg cells from LP (n = 2), PPs (n = 3) and spleens (n = 3). DE analysis was performed using limma. P values were adjusted to control the global FDR across all comparisons with the ‘global’ option of the limma package. (d) Gene set enrichment plots showing significantly enriched (FDR < 0.1) hallmark gene sets in c-Maf-deficient versus control LP Treg cells. Data represent the combined analysis of two biologically independent samples. (e) MA plot showing comparison of gene expression between LP (n = 2) and splenic Treg cells (n = 3) from control mice. Genes up-regulated in LP Treg cells are highlighted in red; genes down-regulated are highlighted in blue (FDR < 0.05). DE analysis was performed using limma. P values were adjusted to control the global FDR across all comparisons with the ‘global’ option of the limma package. (f) Sorted c-Maf-deficient and control LP Treg cells (n = 3) were subjected to ‘Assay for Transposase-Accessible Chromatin’ sequencing (ATAC-seq). MA plot displaying regions of accessible chromatin. Regions over-represented in c-Maf-deficient Treg cells are highlighted in red; regions under-represented are highlighted in blue. Note that multiple peaks can be associated with individual genes. (g) Representative c-Maf chromatin immunoprecipitation sequencing (ChIP-seq) and ATAC sequencing tracks. Arrows indicate c-Maf occupancy in open chromatin regions of Treg cell gene loci. Data represent the combined analysis of three biologically independent samples. c-Maf ChIP-seq was published by Ciofani et al.34
Supplementary Figure 6 Il10∆Treg mice are susceptible to DSS-induced colitis.
Foxp3Cre (Ctrl) and Il10fl/flFoxp3Cre (Il10∆Treg) mice were orally challenged with 1.5% DSS (w/v) for 5 days. Colitis severity was assessed 3 days later (day 8). (a) Colon length (n = 4, mean ± SEM) and (b) colitis scores are shown (n = 4, mean ± SEM). *P = 0.03 as calculated by unpaired Student’s t-test (two-tailed). Data are representative of three independent experiments with four to five mice per genotype.
Supplementary Figure 7 c-Maf is essential for Treg cell–mediated control of intestinal IgA responses.
Analysis of faecal microbiota diversity of co-housed littermate Maf∆Treg (n = 10, mean ± SEM) and control mice (n = 11, mean ± SEM) using 16 S rRNA gene sequencing. (b) Quantification of IgA in serum of Maf∆Treg and control mice as measured by ELISA (n = 4, mean ± SEM, *P = 0.0129). (c) Frequency of IgA+ B cells in PP of naïve Maf∆Treg and control mice. Representative contour plots of IgA versus B220 expression by CD45+ cells are shown on the left. The graph on the right shows quantification (n = 4, mean ± SEM). (d) Quantification of IgA+ cells in the colonic LP of Maf∆Treg and control mice as measured by immunohistochemistry (n = 4, mean ± SEM, *P = 0.0045). (e) Differential representation of bacterial families between the IgA+ and IgA− fractions of co-housed littermate Maf∆Treg and control mice shown in form of an IgA coating index (see Methods section). For a given taxon, the value of the IgA coating index can range from a maximum of 1.0 (taxon detected exclusively in the IgA+ fraction) to a minimum of −1.0 (present only in the IgA− fraction). n = 4, *P = 0.0286 as calculated by Mann–Whitney U test (two-tailed). Whiskers represent the minimum and maximum of data. (f) Quantification of IgA in faeces of Il10∆Treg and control mice as measured by ELISA (n = 4, mean ± SEM). All statistical differences were tested using an unpaired Student’s t-test (two-tailed) unless stated otherwise.
Supplementary Figure 8 Phenotypic analysis of intestinal Foxp3+ Treg cells in colonized germ-free mice.
(a) Experimental design for colonization of germ-free (GF) mice with microbiota (Mb) from either Maf∆Treg or control mice. (b–d) GF + Ctrl Mb (n = 6, mean ± SEM) and GF + Maf∆Treg Mb (n = 8, mean ± SEM) mice were orally challenged with 1.5% DSS (w/v) for 5 days. Colonic Foxp3+ Treg cells were analyzed 2 days later. Graphs show pooled data of two independent experiments. (b) Frequency of Foxp3+ Treg cells among colonic CD4+ TCRβ+ T cells. (c) Frequency of c-Maf+ and RORγt+ cells among colonic Foxp3+ Treg cells. (d) Frequency of IL-10+ cells among colonic Foxp3+ Treg cells after ex vivo PMA/ionomycin restimulation. (e) Frequency of TFR cells (PD1+ CXCR5+) among Foxp3+ cells from PPs of GF + Ctrl Mb (n = 4, mean ± SEM) and GF + Maf∆Treg Mb (n = 3, mean ± SEM) mice after 2 weeks of co-housing with either Maf∆Treg or control mice. (f) Quantification of IgA in faeces of GF + Ctrl Mb (n = 4, mean ± SEM) and GF + Maf∆Treg Mb (n = 3, mean ± SEM) mice after 2 weeks of co-housing with either Maf∆Treg or control mice as measured by ELISA. (e–f) Data are representative of two independent experiments.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1–5
Rights and permissions
About this article
Cite this article
Neumann, C., Blume, J., Roy, U. et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host–microbiota homeostasis. Nat Immunol 20, 471–481 (2019). https://doi.org/10.1038/s41590-019-0316-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0316-2
This article is cited by
-
Blimp-1 and c-Maf regulate immune gene networks to protect against distinct pathways of pathobiont-induced colitis
Nature Immunology (2024)
-
Strategies for targeting cytokines in inflammatory bowel disease
Nature Reviews Immunology (2024)
-
Single-cell transcriptomic analysis of normal and pathological tissues from the same patient uncovers colon cancer progression
Cell & Bioscience (2023)
-
The role of transcription factors in shaping regulatory T cell identity
Nature Reviews Immunology (2023)
-
Regulatory T cells in the face of the intestinal microbiota
Nature Reviews Immunology (2023)