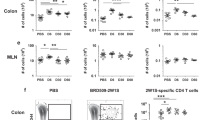
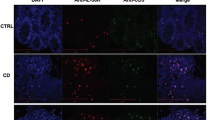
Abstract
Although tissue-resident memory T cells (TRM cells) have been shown to regulate host protection in infectious disorders, their function in inflammatory bowel disease (IBD) remains to be investigated. Here we characterized TRM cells in human IBD and in experimental models of intestinal inflammation. Pro-inflammatory TRM cells accumulated in the mucosa of patients with IBD, and the presence of CD4+CD69+CD103+ TRM cells was predictive of the development of flares. In vivo, functional impairment of TRM cells in mice with double knockout of the TRM-cell-associated transcription factors Hobit and Blimp-1 attenuated disease in several models of colitis, due to impaired cross-talk between the adaptive and innate immune system. Finally, depletion of TRM cells led to a suppression of colitis activity. Together, our data demonstrate a central role for TRM cells in the pathogenesis of chronic intestinal inflammation and suggest that these cells could be targets for future therapeutic approaches in IBD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
07 March 2019
In the version of this article initially published, a portion of the Acknowledgements section (“the Clinical Research Group CEDER of the German Research Council (DFG)”) was incorrect. The correct statement is as follows: “...the Collaborative Research Center TRR241 of the German Research Council (DFG)...”. The error has been corrected in the HTML and PDF version of the article.
References
Baumgart, D. C. & Sandborn, W. J. Crohn’s disease. Lancet 380, 1590–1605 (2012).
Strober, W., Fuss, I. & Mannon, P. The fundamental basis of inflammatory bowel disease. J. Clin. Invest. 117, 514–521 (2007).
Macdonald, T. T. & Monteleone, G. Immunity, inflammation, and allergy in the gut. Science 307, 1920–1925 (2005).
Kaser, A., Zeissig, S. & Blumberg, R. S. Inflammatory bowel disease. Annu. Rev. Immunol. 28, 573–621 (2010).
Neurath, M. F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14, 329–342 (2014).
Lee, J. C. et al. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat. Genet. 49, 262–268 (2017).
Ananthakrishnan, A. N. et al. Environmental triggers in IBD: a review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol. 15, 39–49 (2018).
Kaser, A. & Tilg, H. Novel therapeutic targets in the treatment of IBD. Expert. Opin. Ther. Targets 12, 553–563 (2008).
Powrie, F. et al. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity 1, 553–562 (1994).
Zundler, S. & Neurath, M. F. Pathogenic T cell subsets in allergic and chronic inflammatory bowel disorders. Immunol. Rev. 278, 263–276 (2017).
Neurath, M. F. et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J. Exp. Med. 195, 1129–1143 (2002).
Heller, F., Fuss, I. J., Nieuwenhuis, E. E., Blumberg, R. S. & Strober, W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity 17, 629–638 (2002).
Gerlach, K. et al. TH9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat. Immunol. 15, 676–686 (2014).
Ahern, P. P. et al. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 33, 279–288 (2010).
Maul, J. et al. Peripheral and intestinal regulatory CD4+CD25high T cells in inflammatory bowel disease. Gastroenterology 128, 1868–1878 (2005).
Feagan, B. G. et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med. 369, 699–710 (2013).
Vermeire, S. et al. Etrolizumab as induction therapy for ulcerative colitis: a randomised, controlled, phase 2 trial. Lancet 384, 309–318 (2014).
Zundler, S. & Neurath, M. F. Novel insights into the mechanisms of gut homing and antiadhesion therapies in inflammatory bowel diseases. Inflamm. Bowel Dis. 23, 617–627 (2017).
Schenkel, J. M., Fraser, K. A., Vezys, V. & Masopust, D. Sensing and alarm function of resident memory CD8+ T cells. Nat. Immunol. 14, 509–513 (2013).
Jiang, X. et al. Skin infection generates non-migratory memory CD8+ TRM cells providing global skin immunity. Nature 483, 227–231 (2012).
Sheridan, B. S. et al. Oral infection drives a distinct population of intestinal resident memory CD8+ T cells with enhanced protective function. Immunity 40, 747–757 (2014).
Gebhardt, T. et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10, 524–530 (2009).
Schenkel, J. M. & Masopust, D. Tissue-resident memory T cells. Immunity 41, 886–897 (2014).
Park, C. O. & Kupper, T. S. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat. Med. 21, 688–697 (2015).
Ariotti, S. et al. T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105 (2014).
Mueller, S. N. & Mackay, L. K. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16, 79–89 (2016).
Mackay, L. K. & Kallies, A. Transcriptional regulation of tissue-resident lymphocytes. Trends Immunol. 38, 94–103 (2017).
Kumar, B. V. et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017).
Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016).
van Gisbergen, K. P. J. M. et al. Mouse Hobit is a homolog of the transcriptional repressor Blimp-1 that regulates NKT cell effector differentiation. Nat. Immunol. 13, 864–871 (2012).
Milner, J. J. et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017).
Hombrink, P. et al. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 17, 1467–1478 (2016).
Skon, C. N. et al. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 14, 1285–1293 (2013).
Stelma, F. et al. Human intrahepatic CD69+CD8+ T cells have a tissue resident memory T cell phenotype with reduced cytolytic capacity. Sci. Rep. 7, 6172 (2017).
Ashouri, J. F. & Weiss, A. Endogenous Nur77 is a specific indicator of antigen receptor signaling in human T and B cells. J. Immunol. 198, 657–668 (2017).
Thom, J. T., Weber, T. C., Walton, S. M., Torti, N. & Oxenius, A. The salivary gland acts as a sink for tissue-resident memory CD8+ T cells, facilitating protection from local cytomegalovirus infection. Cell Rep. 13, 1125–1136 (2015).
Teijaro, J. R. et al. Cutting edge: tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187, 5510–5514 (2011).
Turner, D. L. & Farber, D. L. Mucosal resident memory CD4 T cells in protection and immunopathology. Front. Immunol. 5, 331 (2014).
Zundler, S. et al. Three-dimensional cross-sectional light-sheet microscopy imaging of the inflamed mouse gut. Gastroenterology 153, 898–900 (2017).
Fischer, A. et al. Differential effects of α4β7 and GPR15 on homing of effector and regulatory T cells from patients with UC to the inflamed gut in vivo. Gut 65, 1642–1664 (2016).
Wirtz, S. et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 12, 1295–1309 (2017).
Fernandez-Ruiz, D. et al. Liver-resident memory CD8+ T cells form a front-line defense against malaria liver-stage infection. Immunity 45, 889–902 (2016).
Hong, S. et al. Differential regulation of P2X7 receptor activation by extracellular nicotinamide adenine dinucleotide and ecto-ADP-ribosyltransferases in murine macrophages and T cells. J. Immunol. 183, 578–592 (2009).
Carta, S. et al. Cell stress increases ATP release in NLRP3 inflammasome-mediated autoinflammatory diseases, resulting in cytokine imbalance. Proc. Natl Acad. Sci. USA 112, 2835–2840 (2015).
Turner, D. L. et al. Biased generation and in situ activation of lung tissue-resident memory CD4 T cells in the pathogenesis of allergic asthma. J. Immunol. 200, 1561–1569 (2018).
Oja, A. E. et al. The transcription factor Hobit identifies human cytotoxic CD4+ T cells. Front. Immunol. 8, 325 (2017).
Vieira Braga, F. A. et al. Blimp-1 homolog Hobit identifies effector-type lymphocytes in humans. Eur. J. Immunol. 45, 2945–2958 (2015).
Strober, W. & Fuss, I. J. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology 140, 1756–1767 (2011).
Zundler, S. et al. Blockade of αEβ7 integrin suppresses accumulation of CD8+ and Th9 lymphocytes from patients with IBD in the inflamed gut in vivo. Gut 66, 1936–1948 (2017).
Becker, C., Fantini, M. C. & Neurath, M. F. High resolution colonoscopy in live mice. Nat. Protoc. 1, 2900–2904 (2006).
Erben, U. et al. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 7, 4557–4576 (2014).
Klingberg, A. et al. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J. Am. Soc. Nephrol. 28, 452–459 (2017).
Ullrich, E. et al. BATF-dependent IL-7RhiGM-CSF+T cells control intestinal graft-versus-host disease. J. Clin. Invest. 128, 916–930 (2018).
López-Posadas, R. et al. Rho-A prenylation and signaling link epithelial homeostasis to intestinal inflammation. J. Clin. Invest. 126, 611–626 (2016).
Acknowledgements
This work was funded by a grant of the German Research foundation to S.Z. (ZU 377/1-1). The research of S.Z., R.A., T.R., M.L., K.H., R.L.-P., C.N., I.A., and M.F.N. was supported by the Interdisciplinary Center for Clinical Research (IZKF) and the ELAN program of the University Erlangen-Nuremberg, the Fritz-Bender-Stiftung, the Else Kröner-Fresenius-Stiftung, the German Crohn’s and Colitis Foundation (DCCV), the Collaborative Research Center TRR241 of the German Research Council (DFG), the DFG topic program on Microbiota, the Emerging Field Initiative, the DFG Collaborative Research Centers 643, 796 and 1181 and the Litwin IBD Pioneers program of the Crohn’s and Colitis Foundation of America (CCFA). The authors thank J. Derdau, D. Dziony, J. Schuster, A. Marosan, and K. Enderle for excellent technical assistance. This work was performed in partial fulfillment of the requirements for obtaining the degree “Dr. rer. biol. hum.” for S.Z.
Author information
Authors and Affiliations
Contributions
S.Z., E.B., M. Spocinska, M. Slawik, and M.W. performed the experiments. S.Z., L.P.-V., R.S., R.A., T.R., M.L., K.H., R.L.-P., C.N., I.A., K.P.J.M.v.G., and M.F.N. provided clinical samples, protocols, or reagents, or designed experiments; A.B.E. and S.L. performed and analyzed RNA sequencing; S.Z., E.B., M. Spocinska, C.N., I.A., K.P.J.M.v.G., and M.F.N. analyzed and interpreted the data; S.Z. drafted the manuscript with the help of K.P.J.M.v.G. and M.F.N.; all of the authors critically revised the manuscript for important intellectual content.
Corresponding author
Ethics declarations
Competing interests
M.F.N. has served as an advisor for Pentax, Giuliani, MSD, Abbvie, Janssen, Takeda and Boehringer. M.F.N. and S.Z. received research support from Takeda and Roche. The other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Characterization of TRM cells in IBD.
(a) Flow cytometry of lamina propria mononuclear cells (LPMCs). Left panels: Representative dot plots indicating the expression of the T cell markers CD4 or CD8 in total LPMCs and CD69+CD103+ LPMCs. Right panels: Quantification of T cells expressing CD4 and/or CD8 among CD69+CD103+ LPMCs in CON (n = 4), UC (n = 3) and CD (n = 8). (b) Correlation of CD69 mRNA with CXCR6 and CD101 mRNA expression as determined by qPCR in intestinal tissue from patients with IBD (n = 35). Spearman’s R and regression lines are indicated, * P < 0.05. (c, e) mRNA expression of CRTAM1 (c, n = 5) and NR4A1 (e, n = 6) in CD3+CD69+ vs. CD3+CD69- T cells isolated from colonic lamina propria as determined by qPCR. Pairwise normalization to the expression in CD3+CD69+ T cells was performed. *** P < 0.001, paired two-tailed student’s t-test. (d) Quantitative flow cytometry of CD44 mean fluorescence intensity (MFI) in LPMC subsets. n = 16, *** P < 0.001, repeated-measures ANOVA with Newman-Keuls post-hoc test. (f) Analysis of TRM-associated molecules on CD69+ TRM cells from the intestinal lamina propria (LP) and T cells from the peripheral blood. Histograms of the expression of CD49a and CXCR6 representative for two independent experiments are shown. (g) Correlation of CD69 mRNA expression with mRNA expression of IFNG (n = 24), IL13 (n = 26), IL17A (n = 31), IL12A (n = 46), IL12B (n = 28) and TNF (n = 36) as determined by qPCR in intestinal tissue from patients with IBD. Spearman’s R and regression lines are indicated; * P < 0.05, ** P < 0.01, *** P < 0.001.
Supplementary Figure 2 CD4+ and CD8+ TRM cells in IBD.
(a) Histograms showing the expression of CD44, S1PR1 (CD363) and CD11a in CD69+ cells from the peripheral blood (PB) and on CD69+CD103- and CD69+CD103+ CD4+ lamina propria mononuclear cells (LPMCs) from endoscopic biopsies as indicated. Data are representative for two independent experiments. (b) Representative gating strategy for human CD4+ and CD8+ CD69+CD103+ TRM cells. From left to right: After exclusion of doublets, lymphocytes were selected in the foreward-/sideward-scatter. CD4+ and CD8+ T cells were identified and CD69 was plotted against CD103 to select CD69+CD103+ TRM cells. (c) The frequency of CD4+ and CD8+ CD69+ TRM cells (left) and total CD4+ and CD8+ T cells (right) in a cohort of 46 IBD patients was determined by flow cytometry and the respective median was calculated. Flare-free survival of patients with frequencies above (high) or below (low) the median was determined. P-values of one-sided log-rank comparison are indicated.
Supplementary Figure 3 Characterization of mouse TRM cells and comparable abundance of CD4+ Hobit–Blimp-1 double-knockout (DKO) and WT T cells and TRM cells in transfer colitis.
(a) Representative gating strategy for mouse CD4+ CD44+CD69+ TRM cells. From left to right: After exclusion of doublets and dead cells, lymphocytes were selected in the foreward-/sideward-scatter. CD3+CD4+ T cells were identified and CD44 was plotted against CD69 to select CD44+CD69+ TRM cells. (b) Flow cytometric quantification of CD44+CD69+CD103+ TRM frequency in LPMCs of Rag1-/- mice after transfer of naïve CD4+ T cells. Left panels: Representative dot plots of CD3+CD4+CD44+ LPMCs one week and six weeks after transfer; the frequency of CD69+CD103+ TRM cells among lymphoid LPMCs is indicated. Right panels: Quantification of CD103+ CD4+ TRM cell frequency over time. n = 5 (1+2 weeks), n = 4 (3+4 weeks), n = 3 (5 weeks). (c) Flow cytometry of the expression of the TRM-associated molecule CD49a on CD44+CD69+ TRM cells from the intestinal lamina propria (LP) and splenic CD44+ memory T cells. Histograms representative for two independent experiments are shown. (d) Immunohistochemistry staining for CD4 in colon tissue of Rag1-/- mice after transfer of naïve CD4+ WT, Hobit KO or DKO cells. Left panels: Representative images. Right panel: Quantitative data. Bars: 50 µm (upper row), 25 µm (lower row). n = 19 from two independent experiments. (e) Left: Competitive in vivo homing assay (n = 8) with CD4+ T cells from DKO (green) and WT (magenta) mice transferred to the ileocolic artery of Rag1-/- mice. Representative intravital confocal microscopy with arrows highlighting extravasated cells and quantification of homed cells per region of interest (ROI). Right: Non-competitive in vivo homing assay (n = 10) with CD4+ T cells from DKO and WT mice transferred to the ileocolic artery of Rag1-/- mice. Representative dot plots of homed CFSE+ cells in the LP and quantification. (f) Flow cytometry assessment of proliferation and apoptosis of anti-CD3/CD28-stimulated CD4+ WT and DKO T cells. Left panels: Representative flow cytometry showing CFSE dilution after culture of CD4+ T cells for 48 hours. Right panels: Quantification of proliferation and apoptosis as indicated. n = 10. (g) Flow cytometry of CD69 and CD103 expression in CD4+ LPMCs from Rag1-/- mice after transfer of WT or DKO cells. Representative histograms and quantitative data from one representative experiment with six mice out of two independent experiments are shown. (b, d) Center values – mean; error bars – s.e.m., (e,f) Boxplots – median with upper and lower quartile, whiskers – minimum and maximum. (g) Floating bars - median with minimum and maximum. (d) One-way ANOVA, (e-g) two-tailed student’s t-test. n.s. – not significant.
Supplementary Figure 4 Pathogenic mechanisms of TRM cells in T cell transfer colitis.
(a) Functional annotation analysis of previously published whole genome transcript analysis in WT and DKO NKT cells (Mackay et al. Science 352, 459–463) with Ingenuity Pathway Analysis (Qiagen). Using the “Diseases and Functions” mode the function “Recruitment” (subordinate to the function “Cell Movement”) was analyzed. Boxes are sized according to the respective -log(p) value and colored according to the z-score (right) assigned to the respective function. Fisher’s Exact test. (b) Expression of Cxcl3 mRNA in LPMCs from Rag1-/- mice transferred with WT, Hobit KO and DKO cells as determined by qPCR. n = 27 from three independent experiments. (c) Correlation of CD69 mRNA expression with mRNA expression of CCL3 (n = 37) and CCL4 (n = 34) as determined by qPCR in intestinal tissue from patients with IBD. Spearman’s R and regression lines are indicated; * P < 0.05. (d) mRNA expression of CCL3 and CCL4 in CD3+CD69+ vs. CD3+CD69- T cells isolated from colon lamina propria as determined by qPCR. Pairwise normalization to the expression in CD3+CD69+ T cells was performed. n = 6. (e, f) Expression of IL-2 in mRNA from LPMCs of RAG1–/– mice after transfer of WT, Hobit KO and DKO cells as determined by qPCR (e) and in supernatants from anti-CD3/CD28-stimulated LPMCs as determined by multiplex assay (n = 20 from two independent experiments). (g) Left panels: mRNA expression of Gzmb and Prf1 in LPMCs from the colon of Rag1-/- mice after transfer of WT, Hobit KO and DKO cells as determined by qPCR (n = 27 from three independent experiments). Right panel: Granzyme B concentration in the supernatants of anti-CD3/CD28-stimulated LPMCs from such mice as determined by ELISA (n = 30 from three independent experiments). (h) Culture of organoids in pure crypt culture medium (CCM) or with 25 % supernatant from WT T cells or from DKO T cells. Left panels: Representative bright-field and fluorescence microscopy images of organoid architecture and dead dye (red)/nuclei dye (blue) intensity, respectively; right panels: quantification of cell death. n = 24. (i) Immunohistochemistry staining for EpCAM (red) and TUNEL (green) in colon tissue from Rag1-/- mice after transfer of WT, Hobit KO or DKO cells. Left: Representative images, white arrows highlight EpCAM+TUNEL+ apoptotic epithelial cells. Right: Quantification of EpCAM+TUNEL+ cells per high power field (HPF). n = 21 from two independent experiments. Bars: 50 µm (upper row), 25 µm (lower row). (d, i) Center values – mean; error bars – s.e.m., (b, e-h) Boxplots – median with upper and lower quartile, whiskers – minimum and maximum. (b, d-i) * P < 0.05, ** P < 0.01, *** P < 0.001, one-way ANOVA with Newman-Keuls post-hoc testing (b, e-g, i) or paired (d) or unpaired (h) two-tailed student’s t-test.
Supplementary Figure 5 Pathogenic mechanisms of TRM cells in acute DSS-induced colitis.
(a) Immunohistochemistry staining for Foxp3 in colon tissue from WT, Hobit KO or DKO mice with acute DSS colitis. Left: Representative images, white arrows highlight positive cells. Right: Quantification of positive cells per high power field (HPF). n = 26 from three independent experiments; Bars: 50 µm (upper row), 25 µm (lower row). (b) Immunhohistochemistry staining for MPO in colon tissue from WT or DKO mice treated with DSS for three days. Left: Representative images, white arrows highlight positive cells. Right: Quantification of positive cells per HPF. n = 10 from two independent experiments. (c-f) Expression of pro-inflammatory cytokines in mRNA from LPMCs from the colon of WT, Hobit KO and DKO mice with acute DSS colitis as determined by qPCR (c, e) and in supernatants from anti-CD3/CD28-stimulated LPMCs as determined by multiplex assay (d, f). n = 17 (c, Il12a, e, Il13), n = 16 (e, Ifng, Il17a), n = 15 (c, Tnf) from two independent experiments, n = 8 from one representative out of two independent experiments (c, Il1b), n = 22 from three independent experiments (d, f). (g) mRNA expression of Gzmb in LPMCs from the colon of WT, Hobit KO and DKO mice with acute DSS colitis as determined by qPCR (left panel, n = 14 from two independent experiments). Granzyme B concentration in the supernatants of anti-CD3/CD28-stimulated LPMCs from such mice as determined by ELISA (right panel, n = 15 from two independent experiments). (h) Immunohistochemistry staining for EpCAM (red) and TUNEL (green) in the colon of WT, Hobit KO and DKO mice with acute DSS colitis Left: Representative images, white arrows highlight EpCAM+TUNEL+ apoptotic epithelial cells. Right: Quantification of EpCAM+TUNEL+ cells per high power field (HPF). n = 19 from two independent experiments. Bars: 50 µm (upper row), 25 µm (lower row). (a, b, h) Center values – mean; error bars – s.e.m.; (c-g) Boxplots – median with upper and lower quartile, whiskers – minimum and maximum; floating bars - median with minimum and maximum. (a-h) * P < 0.05, ** P < 0.01, *** P < 0.001, two-tailed student’s t-test (b, d IL-12, e Il17a, f IL-13), one-way ANOVA with Newman-Keuls post hoc test (all other panels).
Supplementary Figure 6 Depletion of TRM cells via DT.
(a, b) Flow cytometry of CD4+ T cell (a) and CD103+ CD4+ TRM cell frequency (b) in LPMCs of Rag1-/- mice after transfer of naive CD4+ Hobit-DTR cells and treatment without (“no DT”) or with early or late diphtheria toxin (DT). (a) Representative histograms for 10 mice from one experiment are shown. (b) Left: representative flow cytometry, the frequency of CD69+CD103+ TRM cells among lymphoid LPMCs is indicated. Right: Quantification of CD69+CD103+ TRM cells among total lymphoid LPMCs; n = 21 from three independent experiments. (c) Immunohistochemistry staining for CD4 (left, n = 24) and MPO (right, n = 22) in colon tissue of Rag1-/- mice after transfer of naive CD4+ Hobit-DTR cells and treatment without or with early and late DT. Representative images from three independent experiments with white arrows highlighting CD4+ or MPO+ cells and quantification of CD4+ or MPO+ cells per high power field (HPF). Bars: 50µm (upper row), 25µm (lower row). (d) mRNA expression of chemokines and pro-inflammatory cytokines in LPMCs from the colon of Rag1-/- mice after transfer of naiveCD4+ Hobit-DTR cells and treatment without or with early and late DT as determined by qPCR. n = 14 (Ccl3, Ccl4, Ifng), n = 13 (Cxcl2, Il17, Il1b), n = 12 (Il12a) from two independent experiments, n = 21 from three independent experiments (Il13). (e) Concentration of pro-inflammatory cytokines in the supernatants of anti-CD3/CD28-stimulated LPMCs from the colon of Rag1-/- mice after transfer of naiveCD4+ Hobit-DTR cells and treatment without or with early and late DT as determined by multiplex assay. n = 10 (IFN-γ, IL-13), n = 9 (IL-17A, IL-1β). = 9–10. (b-d) Boxplots – median with upper and lower quartile, whiskers – minimum and maximum. (e) Floating bars - median with minimum and maximum. * P < 0.05, ** P < 0.01, *** P < 0.001, two-tailed student’s t-test (d, Il13, Il17a) or one-way ANOVA with Newman-Keuls post-hoc comparison (all other panels).
Supplementary Figure 7 Depletion of TRM cells via NAD.
(a) Flow cytometry of P2X7-Art2.2-coexpression. Representative control stainings for Fig. 7f showing the expression of P2X7 and Art2.2 in spleen CD3- and CD3+ cells. The frequency of double-positive cells is indicated. (b) Frequency of CD44+CD69- effector memory T cells (TEM) in mice treated with or without NAD. n = 22. (c) Representative (left) and quantitative (right) flow cytometry of CD1d tetramer+ NKT cells in the colon of mice treated with or without NAD as well as representative gating control flow cytometry of CD1d tetramer+ NKT cells in the liver (far right). n = 14. (d) Frequency of CD3+ T and CD1d tetramer+ NKT cells among lymphoid LPMCs after treatment with PBS. n = 8. (e) Immunohistochemistry staining for CD4 (n = 16) and MPO (n = 17) in colon tissue of WT mice with TNBS colitis after treatment with or without NAD from three independent experiments. (f) Flow cytometry of Art2.2 expression in T cells versus non-T cells. Left: Representative histograms of Art2.2 expression on CD4+ T cells, CD3- non T cells and fluorescence minus one (FMO) control for Art2.2. Right: Quantitative expression. n = 10 mice from three independent experiments. (b-f) Boxplots – median with upper and lower quartile, whiskers – min and max. n.s. – not significant, * P < 0.05, ** P < 0.01, *** P < 0.001, paired (d, f) or unpaired (b, c, e) two-tailed student’s t-test.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–3
Supplementary Video 1: Light-sheet fluorescence microscopy
Representative animation of a colon sample from a Rag1–/– mouse after transfer of naïve CD4+ T cells from a Hobit–Blimp-1 double-knockout mouse. Whole-mount staining for CD4 (red) and autofluorescence signal (grey) were recorded by light-sheet fluorescence microscopy
Rights and permissions
About this article
Cite this article
Zundler, S., Becker, E., Spocinska, M. et al. Hobit- and Blimp-1-driven CD4+ tissue-resident memory T cells control chronic intestinal inflammation. Nat Immunol 20, 288–300 (2019). https://doi.org/10.1038/s41590-018-0298-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-018-0298-5
This article is cited by
-
Activation of mucosal insulin receptor exacerbates intestinal inflammation by promoting tissue resident memory T cells differentiation through EZH2
Journal of Translational Medicine (2024)
-
Targeting integrin pathways: mechanisms and advances in therapy
Signal Transduction and Targeted Therapy (2023)
-
Gut immune cell trafficking: inter-organ communication and immune-mediated inflammation
Nature Reviews Gastroenterology & Hepatology (2023)
-
Cancer-specific tissue-resident memory T-cells express ZNF683 in colorectal cancer
British Journal of Cancer (2023)
-
CD4+ T cell memory
Nature Immunology (2023)