Abstract
Germinal center (GC) B cells feature repression of many gene enhancers to establish their characteristic transcriptome. Here we show that conditional deletion of Lsd1 in GCs significantly impaired GC formation, associated with failure to repress immune synapse genes linked to GC exit, which are also direct targets of the transcriptional repressor BCL6. We found that BCL6 directly binds LSD1 and recruits it primarily to intergenic and intronic enhancers. Conditional deletion of Lsd1 suppressed GC hyperplasia caused by constitutive expression of BCL6 and significantly delayed BCL6-driven lymphomagenesis. Administration of catalytic inhibitors of LSD1 had little effect on GC formation or GC-derived lymphoma cells. Using a CRISPR-Cas9 domain screen, we found instead that the LSD1 Tower domain was critical for dependence on LSD1 in GC-derived B cells. These results indicate an essential role for LSD1 in the humoral immune response, where it modulates enhancer function by forming repression complexes with BCL6.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Sequence data that support the findings of this study have been deposited in GEO under SuperSeries GSE118783 with the accession codes GSE106417 (RNA-seq for GC B cell), GSE106416 (RNA-seq for lymphoma) and GSE107920 (ChIP-seq). Other data that support these findings are available from the corresponding author upon reasonable request.
References
Mesin, L., Ersching, J. & Victora, G. D. Germinal center B cell dynamics. Immunity 45, 471–482 (2016).
Hatzi, K. & Melnick, A. Breaking bad in the germinal center: how deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 20, 343–352 (2014).
Beguelin, W. et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013).
Beguelin, W. et al. EZH2 and BCL6 cooperate to assemble CBX8-BCOR complex to repress bivalent promoters, mediate germinal center formation and lymphomagenesis. Cancer Cell 30, 197–213 (2016).
Bunting, K. L. et al. Multi-tiered reorganization of the genome during B cell affinity maturation anchored by a germinal center-specific locus control region. Immunity 45, 497–512 (2016).
Hatzi, K. et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 4, 578–588 (2013).
Jiang, Y. et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov. 7, 38–53 (2017).
Ortega-Molina, A. et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med. 21, 1199–1208 (2015).
Zhang, J. et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med. 21, 1190–1198 (2015).
Zhang, J. et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 7, 322–337 (2017).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004).
Foster, C. T. et al. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol. Cell. Biol. 30, 4851–4863 (2010).
Wang, J. et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 (2009).
Wang, J. et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446, 882–887 (2007).
Kerenyi, M. A. et al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife 2, e00633 (2013).
Schenk, T. et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat. Med. 18, 605–611 (2012).
Good-Jacobson, K. L., Song, E., Anderson, S., Sharpe, A. H. & Shlomchik, M. J. CD80 expression on B cells regulates murine T follicular helper development, germinal center B cell survival, and plasma cell generation. J. Immunol. 188, 4217–4225 (2012).
Piovesan, D. et al. c-Myb regulates the T-Bet-dependent differentiation program in b cells to coordinate antibody responses. Cell Rep. 19, 461–470 (2017).
Kim, S. J. et al. Increased IL-12 inhibits B cells’ differentiation to germinal center cells and promotes differentiation to short-lived plasmablasts. J. Exp. Med. 205, 2437–2448 (2008).
Bond, J. et al. Direct interaction of Ikaros and Foxp1 modulates expression of the G protein-coupled receptor G2A in B-lymphocytes and acute lymphoblastic leukemia. Oncotarget 7, 65923–65936 (2016).
Huang, C. et al. The BCL6 RD2 domain governs commitment of activated B cells to form germinal centers. Cell Rep. 8, 1497–1508 (2014).
Mohammad, H. P. et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 28, 57–69 (2015).
Mohammad, H. et al. Inhibition of LSD1 as a therapeutic strategy for the treatment of acute myeloid leukemia. Blood 122, 3964–3964 (2013).
Sprussel, A. et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia 26, 2039–2051 (2012).
Shi, J. et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 (2015).
Stavropoulos, P., Blobel, G. & Hoelz, A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat. Struct. Mol. Biol. 13, 626–632 (2006).
Chen, Y. et al. Crystal structure of human histone lysine-specific demethylase 1 (LSD1). Proc. Natl Acad. Sci. USA 103, 13956–13961 (2006).
Forneris, F., Binda, C., Adamo, A., Battaglioli, E. & Mattevi, A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J. Biol. Chem. 282, 20070–20074 (2007).
Cerchietti, L. C. et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell 17, 400–411 (2010).
Polo, J. M. et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. 10, 1329–1335 (2004).
Monti, S. et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 105, 1851–1861 (2005).
Cattoretti, G. et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell 7, 445–455 (2005).
Shi, Y. J. et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864 (2005).
Yang, M. et al. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell 23, 377–387 (2006).
Karytinos, A. et al. A novel mammalian flavin-dependent histone demethylase. J. Biol. Chem. 284, 17775–17782 (2009).
Harris, W. J. et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21, 473–487 (2012).
Cusan, M. et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1- and C/EBPalpha-dependent enhancers in AML. Blood 131, 1730–1742 (2018).
Stamos, D., Li, L., Rosenfeld, M. & Love, P. E. A requirement for lysine-specific histone demethylase 1A (Lsd1) for T cell development. J. Immunol. 200, 165.24 (2018).
Su, S. T. et al. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol. Cell. Biol. 29, 1421–1431 (2009).
Ersching, J. et al. Germinal center selection and affinity maturation require dynamic regulation of mTORC1 Kinase. Immunity 46, 1045–1058.e1046 (2017).
Landt, S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome. Biol. 9, R137 (2008).
Egan, B. et al. An alternative approach to ChIP-Seq normalization enables detection of genome-wide changes in histone H3 Lysine 27 trimethylation upon EZH2 Inhibition. PLoS One 11, e0166438 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 15, 550 (2014).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Lun, A. T. & Smyth, G. K. De novo detection of differentially bound regions for ChIP-seq data using peaks and windows: controlling error rates correctly. Nucleic Acids Res. 42, e95 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Geng, H. et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2, 1004–1023 (2012).
Yoav Benjamini, Y. H. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. 57, 289–300 (1995).
Shaffer, A. L. et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol. Rev. 210, 67–85 (2006).
Acknowledgements
We thank the members of the Melnick and Lowe laboratories for helpful discussions throughout the duration of the project. We also thank T. Hoshii and S. Armstrong for sharing CRISPR-Cas9 reagents and protocols and M. Luo, B. Laurent and Y. Shi for helpful suggestions on LSD1 demethylation assays. A.M.M. is supported by the Chemotherapy Foundation, LLS-TRP 6457-15, LLS SCOR 7012-16, NCI R01 CA187109, NCI R35 CA220499 and the Follicular Lymphoma Consortium. K.H. was supported by the Charles H. Revson Senior Fellowship in Biomedical Science and the NIH/NCI K99CA212276 award.
Author information
Authors and Affiliations
Contributions
K.H. conceptualized the study, designed and performed the experiments, analyzed the data and wrote the manuscript. H.G., A.S.D. and C.M. performed bioinfomatic analyses. R.L., M.C., C.D., H.S., M.N.C.V., T.B. and R.S. assisted in experiments. H.P.M. and R.G.K. provided GSK-LSD1 inhibitor and technical advice regarding drug treatments. A.M.H. edited the manuscript and provided technical advice with flow cytometry and IHC. G.I. performed pathological evaluation of transplanted mice. S.W.L. co-supervised the study. A.M.M. conceptualized and supervised the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.M.M. received research funding from GlaxoSmithKline. H.P.M. and R.G.K. are GlaxoSmithKline employees. R.S. is currently employed by Cancer Genetics Incorporated.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
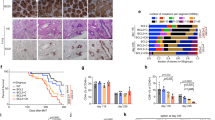
Supplementary Figure 1 LSD1 deletion impairs GC formation.
a) RNA-seq analysis showing levels of other lysine demethylases in human and mouse NB vs GC B cells visualized by heat map FPKM z-score. Data from biologically independent human samples (n = 4 NB, n = 4 GC B) or mouse RNA-seq (n = 6 NB, n = 5 GC B samples). b) Fold BCL6 and AICDA mRNA abundance (mean +/− s.d.) in NB and GC B cells isolated from three biologically independent reactive human tonsils (T1, T2, T3) normalized to HPRT (triplicate qPCR reactions). P value was calculated by two-way ANOVA. c) LSD1 IHC staining of human tonsils (LZ: Light Zone, DZ: Dark Zone, CC: Centrocytes, CB: Centroblasts). Scale bar, 50 μm d) LSD1 mRNA abundance in sorted human NB, GC B cells, plasma (PC) and memory B cells (MC) from RNA-seq (NB n = 4; GCB n = 4; PC n = 3; MC n = 3) and published microarray data (NB n = 5; CB n = 5; CC n = 5; PC n = 5; MC n = 5) (GSE12453). e) B220 staining of spleens from immunized Lsd1fl/fl, Cγ1-Cre Lsd1fl/+ or Cγ1-Cre Lsd1fl/fl mice splenic sections corresponding to Fig. 1d. Images representative of at least 10 biologically independent spleens per group. f) GC counts (mean +/− s.d.) per spleen section. Data from Lsd1fl/fl (n = 10), Cγ1-Cre Lsd1fl/+(n = 14) or Cγ1-Cre Lsd1fl/fl (n = 15) biologically independent mice per group. g) qPCR for intron 1 (non-targeted) and exon 5 (Cre-targeted) on DNA from sorted GC B cells (Fas+GL7+) from immunized Lsd1fl/fl (n = 3) or Cγ1-Cre Lsd1fl/fl (n = 2) biologically independent mice. Data are from duplicate qPCRs. h,i) Flow cytometry analysis of mouse GC B cell splenocytes (n = 5 biologically independent mice per group) immunized with SRBC at day 0 and sacrificed at day 4, 6, 8 and 10 post immunization. GC B cells were identified either as Fas+CD38loB220+ populations or as Fas+GL7+ B220+ populations. j) Comparison of circulating high affinity NP-specific IgG1 antibody titers (NP7-BSA) from Lsd1fl/fl (n = 10 biologically independent mice) versus Cγ1-Cre Lsd1fl/fl (n = 9 biologically independent mice). k) Comparison of affinity maturation (ratio of high to low affinity; anti-NP7 /NP30) for IgG1 or IgG2b and IgG3 isotypes from Lsd1fl/fl versus Cγ1-Cre Lsd1fl/fl mice (n = 16 biologically independent mice per group). l) Comparison by ELISpot of the abundance of high affinity anti-NP IgG1 secreting splenocytes or bone marrow cells from immunized Lsd1fl/fl versus Cγ1-Cre Lsd1fl/fl (n = 6 biologically independent mice). Boxplots are shown as median, interquartile range, and minimum to maximum (d,h,i,k,l). P values calculated by two-sided unpaired t-test (f,h,I,j,k,l) (*P < 0.05, **P < 0.01, ***P < 0.001).
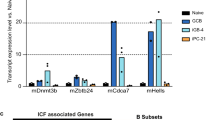
Supplementary Figure 2 B cell–specific LSD1 deletion impairs GC formation without spontaneous differentiation to plasma cell fate.
a) Immunohistochemistry of spleen sections from B cell conditional Lsd1 wild-type (Lsd1fl/fl) or Lsd1-deficient mice (Cd19-Cre Lsd1fl/fl) immunized with SRBC and sacrificed 10 d post immunization stained with PNA at two magnifications (Scale bars, 0.5 mm and 50 μm). b) Quantification of splenic GC area (PNA positive) versus the total area of the spleen section and GC counts per spleen section (n = 3 biologically independent mice per group, mean +/− s.d.) c) Flow cytometry analysis of splenic GC B cells (Fas+CD38lo) within live B cells (B220+DAPI–) from biologically independent Lsd1fl/fl (n = 13) or Cd19-Cre Lsd1fl/fl (n = 12) mice. d) Flow cytometry analysis of splenic GC B cells (Fas+GL7+) within live B cells (B220+DAPI–) from biologically independent Lsd1fl/fl (n = 13) or Cd19-Cre Lsd1fl/fl (n = 13) mice; boxplots with median, interquartile range, and minimum to maximum (c,d). e) Representative flow cytometry dotplots of corresponding GC B cell populations (CD38loFas+ or GL7+Fas+) within live B cells (B220+DAPI–) for Lsd1fl/fl versus Cd19-Cre Lsd1fl/fl mice (c,d). f) Flow cytometry analysis of plasmablasts from Cd19-Cre Lsd1fl/fl versus Lsd1fl/fl littermate mice with at day 6 post immunization with SRBC, stained for cytoplasmic κhi murine splenocytes (cIgκhi) and surface κ (sIgκ). Plasmablasts mostly stained negative for B220 and positive for CD138. g) Flow cytometry analysis of CD138+B220– murine splenocytes representing plasmacytes. P values indicate comparison to Lsd1fl/fl mice by two-sided unpaired t-test (c,d,f,g) (***P < 0.001).
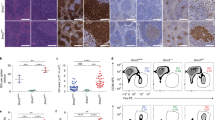
Supplementary Figure 3 LSD1 interacts with BCL6 in GC B cell–derived lymphoma.
a) H3 and H3K4me1 immunoblot of histone demethylation reactions (cropped image) from LSD1, BCL6 and control actin precipitates from lymphoma B cell extracts and bulk core histone substrates with densitometry analysis of band intensity using ImageJ. Representative of two experiments with similar results. b) Coomassie stain and complete BCL6 western blot of fluorography SDS PAGE shown in Fig. 4c. Experiment was performed at least twice with similar result. c) H3K4me1 signal in control regions (50 kb upstream of BCL6-LSD1 binding sites) from B cells transduced with control siRNA or BCL6 targeted siRNA. d) ChIP-seq read density heatmaps showing LSD1 and BCL6 binding at regions co-occupied by both proteins in SUDHL4 cells. H3K4me1 ChIP-seq was performed in SUDHL4 treated with siRNA against BCL6 or control siRNA for 24 h. Heatmaps were centered surrounding LSD1 peak summit and were separated in promoter-bound and intergenic/intronic bound regions. Negative control regions 50kb upstream of binding sites are shown as controls. e) H3K4me2 qChIP from SUDHL4 cells treated with siRNA against BCL6 or control siRNA for 24 h (mean +/− s.d.). BCL6-LSD1 co-occupied regions were amplified and a negative control region served as unbound control. P values indicate comparison to siNT by two-sided unpaired t-test. (*P < 0.05, **P < 0.01).
Supplementary Figure 4 Effects of LSD1 depletion in lymphoma cells and validation of LSD1 locus editing.
a,b) Flow cytometry analysis of mature bone marrow granulocytes (Gr1hiCd11b+) (n = 6 biologically independent mice per group; mean +/− s.d.) with two representative dotplots of BM granulocytes from mice treated with GSK-LSD1 or saline control. P value was calculated using two-sided unpaired t-test. c) GSK-LSD1 inhibitor treatment of a panel of lymphoma cell lines along with a leukemia cell line (Kasumi) as positive control. Cell growth was detected by CellTiter-Glo (mean of three wells) 5 days post treatment. The y-axis shows percentage of GSK-LSD1 inhibitor treated cells compared to vehicle. Experiment was repeated more that 3 times with similar results, d) H3K4me2 qChIP using OCI-Ly1 cells treated with 10, 100 and 1000 nM GSK-LSD1 inhibitor or vehicle for 24h. Several LSD1 target regions were amplified along with an unbound locus control (mean +/− s.d.). BCL6-LSD1 co-occupied regions were amplified and a negative control region served as unbound control. e) qPCR for LSD1 gene targets after treatment with GSK-LSD1 10 μM or shRNA mediated LSD1 knockdown at day 5 post puromycin selection (mean +/− s.d.). f) Viable cell counts of DLBCL cells transduced with shRNAs for LSD1 versus control shRNA from biologically independent experiments at day 5 post puromycin selection (mean +/− s.d.). g) BrdU incorporation assay for DLBCL cell lines transduced with control shRNA or two shRNAs targeting LSD1. Cell cycle stage is indicated by corresponding gates. Data were replicated twice with similar results. h) Fold depletion of % GFP+ cells (mean +/− s.d.) bearing sgRNAs targeting different parts of the LSD1 ORF at d40 post doxycycline induction versus d0 based on OCI-Ly1 Cas9 expressing clone 2 (Fig. 5c). i) T7 endonuclease assay was performed on PCR products amplified from the LSD1 locus encoding the Tower or AOD domain targeted by independent sgRNAs. Genomic DNA from cells targeted with control vector served as negative control for the endonuclease reaction. Undigested PCR products are shown next to endonuclease treated for comparison. Data are representative of two experiments with similar results.
Supplementary Figure 5 LSD1 loss of function rescues BCL6-driven GC hyperplasia and lymphomagenesis.
a) B220 flow cytometry staining (mean +/− s.d.) of spleens corresponding to Fig. 6d,f from Lsd1fl/fl mice (n = 8), IμBcl6 Cγ1-Cre Lsd1+/+ mice (n = 11), IμBcl6 Cγ1-Cre Lsd1fl/+ mice (n = 10), IμBcl6 Cγ1-Cre Lsd1fl/fl mice (n = 9), and Cγ1-Cre Lsd1fl/fl mice (n = 3). b) Quantification of PNA staining (mean +/− s.d.) showing the GC numbers per spleen section from Lsd1fl/fl mice (n = 8), IμBcl6 Cγ1-Cre Lsd1+/+ mice (n = 11), IμBcl6 Cγ1-Cre Lsd1fl/+ mice (c10), IμBcl6 Cγ1-Cre Lsd1fl/fl mice (n = 9), and Cγ1-Cre Lsd1fl/fl mice (n = 3). c) Flow cytometry analysis showing the fraction of BrdU+ GC B cells (as % of total B cells) for Lsd1fl/fl mice (n = 7), IμBcl6 Cγ1-Cre Lsd1+/+ mice (n = 10), IμBcl6 Cγ1-Cre Lsd1fl/+ mice (n = 10), IμBcl6 Cγ1-Cre Lsd1fl/fl mice (n = 9), and Cγ1-Cre Lsd1fl/fl mice (n = 4) (median, interquartile range, and minimum to maximum) All P values were calculated using two-sided unpaired t-test (b,c). Each sample represents an individual mouse (a-c). d) Representative immunohistochemistry images of spleen and liver sections from biologically independent mice transplanted with bone marrow from IμBcl6 Cγ1-Cre Lsd1+/+ (n = 18), IμBcl6 Cγ1-Cre Lsd1fl/fl mice (n = 13), Cγ1-Cre mice (n = 10) and Cγ1-Cre Lsd1fl/fl mice (n = 7). Staining with B220 and H&E is shown in two magnifications for spleen and liver sections (scale bars, 500 μm and 20 μm).
Supplementary Figure 6 Examples of flow cytometry gating strategy.
Gating strategy to identify GC B cell populations (a,b) plasmacytes and plasmablasts (c) and mature granulocytes (d).
Supplementary information
Supplementary Figures 1-6
Supplementary Figures 1-6 Supplementary Tables 1 and 2
Supplementary Data Set 1
Murine GC B geneset containing all genes significantly induced in LSD1 knockout GC B cells versus wild-type
Supplementary Data Set 2
Genomic loci significantly gaining or losing accessibility in Lsd1 knockout GC B cells vs wild-type by ATAC-seq
Supplementary Data Set 3
Pathway analysis on Lsd1 repressed GC B gene sets by ATAC and RNA-seq
Supplementary Data Set 4
RNA-seq differential gene expression analysis by DESeq2 for shLSD1 (2 different hairpins versus control)
Rights and permissions
About this article
Cite this article
Hatzi, K., Geng, H., Doane, A.S. et al. Histone demethylase LSD1 is required for germinal center formation and BCL6-driven lymphomagenesis. Nat Immunol 20, 86–96 (2019). https://doi.org/10.1038/s41590-018-0273-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-018-0273-1
This article is cited by
-
MCT1-governed pyruvate metabolism is essential for antibody class-switch recombination through H3K27 acetylation
Nature Communications (2024)
-
LSD1: an emerging face in altering the tumor microenvironment and enhancing immune checkpoint therapy
Journal of Biomedical Science (2023)
-
Histone demethylases in the regulation of immunity and inflammation
Cell Death Discovery (2023)
-
Viscosalactone B, a natural LSD1 inhibitor, inhibits proliferation in vitro and in vivo against prostate cancer cells
Investigational New Drugs (2023)
-
Frequent ZNF217 mutations lead to transcriptional deregulation of interferon signal transduction via altered chromatin accessibility in B cell lymphoma
Leukemia (2023)