Abstract
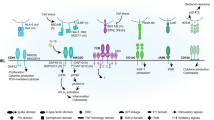
T lymphocytes expressing γδ T cell antigen receptors (TCRs) comprise evolutionarily conserved cells with paradoxical features. On the one hand, clonally expanded γδ T cells with unique specificities typify adaptive immunity. Conversely, large compartments of γδTCR+ intraepithelial lymphocytes (γδ IELs) exhibit limited TCR diversity and effect rapid, innate-like tissue surveillance. The development of several γδ IEL compartments depends on epithelial expression of genes encoding butyrophilin-like (Btnl (mouse) or BTNL (human)) members of the B7 superfamily of T cell co-stimulators. Here we found that responsiveness to Btnl or BTNL proteins was mediated by germline-encoded motifs within the cognate TCR variable γ-chains (Vγ chains) of mouse and human γδ IELs. This was in contrast to diverse antigen recognition by clonally restricted complementarity-determining regions CDR1–CDR3 of the same γδTCRs. Hence, the γδTCR intrinsically combines innate immunity and adaptive immunity by using spatially distinct regions to discriminate non-clonal agonist-selecting elements from clone-specific ligands. The broader implications for antigen-receptor biology are considered.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
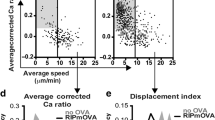
This work did not include any data with mandated deposition in public databases. Associated raw data are provided in the main and/or supplementary figures. Relations to summary data charts are indicated and a full list of figures with associated raw data is provided in the Reporting Summary linked to this article.
References
Hayday, A. C. γδ cells: a right time and a right place for a conserved third way of protection. Annu. Rev. Immunol. 18, 975–1026 (2000).
Davey, M. S. et al. The human Vδ2+ T-cell compartment comprises distinct innate-like Vγ9+ and adaptive Vγ9– subsets. Nat. Commun. 9, 1760 (2018).
Davey, M. S. et al. Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR-dependent adaptive immune surveillance. Nat. Commun. 8, 14760 (2017).
Ravens, S. et al. Human γδ T cells are quickly reconstituted after stem-cell transplantation and show adaptive clonal expansion in response to viral infection. Nat. Immunol. 18, 393–401 (2017).
Gober, H. J. et al. Human T cell receptor γδ cells recognize endogenous mevalonate metabolites in tumor cells. J. Exp. Med. 197, 163–168 (2003).
Adams, E. J., Chien, Y. H. & Garcia, K. C. Structure of a γδ T cell receptor in complex with the nonclassical MHC T22. Science 308, 227–231 (2005).
Luoma, A. M. et al. Crystal structure of Vδ1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human γδ T cells. Immunity 39, 1032–1042 (2013).
Willcox, C. R. et al. Cytomegalovirus and tumor stress surveillance by binding of a human γδ T cell antigen receptor to endothelial protein C receptor. Nat. Immunol. 13, 872–879 (2012).
Groh, V., Steinle, A., Bauer, S. & Spies, T. Recognition of stress-induced MHC molecules by intestinal epithelial γδ T cells. Science 279, 1737–1740 (1998).
Marlin, R. et al. Sensing of cell stress by human γδ TCR-dependent recognition of annexin A2. Proc. Natl. Acad. Sci. USA 114, 3163–3168 (2017).
Bruder, J. et al. Target specificity of an autoreactive pathogenic human γδ-T cell receptor in myositis. J. Biol. Chem. 287, 20986–20995 (2012).
Asarnow, D. M. et al. Limited diversity of γδ antigen receptor genes of Thy-1+ dendritic epidermal cells. Cell 55, 837–847 (1988).
Havran, W. L., Carbone, A. & Allison, J. P. Murine T cells with invariant γδ antigen receptors: origin, repertoire, and specificity. Semin. Immunol. 3, 89–97 (1991).
Kyes, S., Carew, E., Carding, S. R., Janeway, C. A. Jr. & Hayday, A. Diversity in T-cell receptor γ gene usage in intestinal epithelium. Proc. Natl. Acad. Sci. USA 86, 5527–5531 (1989).
Hayday, A. C. Gammadelta T cells and the lymphoid stress-surveillance response. Immunity 31, 184–196 (2009).
Medzhitov, R. & Janeway, C. A. Jr. Innate immunity: the virtues of a nonclonal system of recognition. Cell 91, 295–298 (1997).
Strid, J. et al. Acute upregulation of an NKG2D ligand promotes rapid reorganization of a local immune compartment with pleiotropic effects on carcinogenesis. Nat. Immunol. 9, 146–154 (2008).
Vantourout, P. & Hayday, A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat. Rev. Immunol. 13, 88–100 (2013).
Leishman, A. J. et al. Precursors of functional MHC class I- or class II-restricted CD8αα+ T cells are positively selected in the thymus by agonist self-peptides. Immunity 16, 355–364 (2002).
Boyden, L. M. et al. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal γδ T cells. Nat. Genet. 40, 656–662 (2008).
Turchinovich, G. & Hayday, A. C. Skint-1 identifies a common molecular mechanism for the development of interferon-γ-secreting versus interleukin-17-secreting γδ T cells. Immunity 35, 59–68 (2011).
Di Marco Barros, R. et al. Epithelia use butyrophilin-like molecules to shape organ-specific γδ T cell compartments. Cell 167, 203–218 (2016).
Vantourout, P. et al. Heteromeric interactions regulate butyrophilin (BTN) and BTN-like molecules governing γδ T cell biology. Proc. Natl. Acad. Sci. USA 115, 1039–1044 (2018).
Salim, M. et al. Characterization of a putative receptor binding surface on Skint-1, a critical determinant of dendritic epidermal T cell selection. J. Biol. Chem. 291, 9310–9321 (2016).
Harly, C. et al. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T-cell subset. Blood 120, 2269–2279 (2012).
Kisielow, J., Tortola, L., Weber, J., Karjalainen, K. & Kopf, M. Evidence for the divergence of innate and adaptive T-cell precursors before commitment to the αβ and γδ lineages. Blood 118, 6591–6600 (2011).
Goodman, T. & Lefrancois, L. Intraepithelial lymphocytes. Anatomical site, not T cell receptor form, dictates phenotype and function. J. Exp. Med. 170, 1569–1581 (1989).
San José, E., Borroto, A., Niedergang, F., Alcover, A. & Alarcón, B. Triggering the TCR complex causes the downregulation of nonengaged receptors by a signal transduction-dependent mechanism. Immunity 12, 161–170 (2000).
Heemskerk, M. H. et al. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood 102, 3530–3540 (2003).
Moran, A. E. et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208, 1279–1289 (2011).
Chancellor, A. et al. CD1b-restricted GEM T cell responses are modulated by Mycobacterium tuberculosis mycolic acid meromycolate chains. Proc. Natl. Acad. Sci. USA 114, E10956–E10964 (2017).
Kabelitz, D. et al. New monoclonal antibody (23D12) recognizing three different Vγ elements of the human γδ T cell receptor. 23D12+ cells comprise a major subpopulation of γδ T cells in postnatal thymus. J. Immunol. 152, 3128–3136 (1994).
Hayes, S. M., Shores, E. W. & Love, P. E. An architectural perspective on signaling by the pre-, αβ and γδ T cell receptors. Immunol. Rev. 191, 28–37 (2003).
Palakodeti, A. et al. The molecular basis for modulation of human Vγ9Vδ2 T cell responses by CD277/butyrophilin-3 (BTN3A)-specific antibodies. J. Biol. Chem. 287, 32780–32790 (2012).
Bates, P. A., Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins 5, 39–46 (2001).
Torchala, M., Moal, I. H., Chaleil, R. A., Fernandez-Recio, J. & Bates, P. A. SwarmDock: a server for flexible protein-protein docking. Bioinformatics 29, 807–809 (2013).
Holness, C. L. & Simmons, D. L. Structural motifs for recognition and adhesion in members of the immunoglobulin superfamily. J. Cell Sci. 107, 2065–2070 (1994).
Green, N. et al. Mutational analysis of MAdCAM-1/α4β7 interactions reveals significant binding determinants in both the first and second immunuglobulin domains. Cell Adhes. Commun. 7, 167–181 (1999).
Jones, E. Y. et al. Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 Å resolution. Nature 373, 539–544 (1995).
Moebius, U., Pallai, P., Harrison, S. C. & Reinherz, E. L. Delineation of an extended surface contact area on human CD4 involved in class II major histocompatibility complex binding. Proc. Natl. Acad. Sci. USA 90, 8259–8263 (1993).
Somoza, C., Driscoll, P. C., Cyster, J. G. & Williams, A. F. Mutational analysis of the CD2/CD58 interaction: the binding site for CD58 lies on one face of the first domain of human CD2. J. Exp. Med. 178, 549–558 (1993).
Myers, D. R., Zikherman, J. & Roose, J. P. Tonic signals: why do lymphocytes bother? Trends Immunol. 38, 844–857 (2017).
Hayday, A. & Vantourout, P. A long-playing CD about the γδ TCR repertoire. Immunity 39, 994–996 (2013).
Komori, H. K. et al. Cutting edge: dendritic epidermal γδ T cell ligands are rapidly and locally expressed by keratinocytes following cutaneous wounding. J. Immunol. 188, 2972–2976 (2012).
Fahl, S. P. et al. Role of a selecting ligand in shaping the murine γδ-TCR repertoire. Proc. Natl. Acad. Sci. USA 115, 1889–1894 (2018).
Wencker, M. et al. Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nat. Immunol. 15, 80–87 (2014).
Sumaria, N., Grandjean, C. L., Silva-Santos, B. & Pennington, D. J. Strong TCRγδ signaling prohibits thymic development of iL-17A-secreting γδ T cells. Cell Reports 19, 2469–2476 (2017).
Cernadas, M. et al. Lysosomal localization of murine CD1d mediated by AP-3 is necessary for NK T cell development. J. Immunol. 171, 4149–4155 (2003).
Chiu, Y. H. et al. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat. Immunol. 3, 55–60 (2002).
Mallevaey, T. et al. T cell receptor CDR2β and CDR3β loops collaborate functionally to shape the iNKT cell repertoire. Immunity 31, 60–71 (2009).
Bueno, C. et al. Bacterial superantigens bypass Lck-dependent T cell receptor signaling by activating a Gα11-dependent, PLC-β-mediated pathway. Immunity 25, 67–78 (2006).
Fields, B. A. et al. Crystal structure of a T-cell receptor β-chain complexed with a superantigen. Nature 384, 188–192 (1996).
Kreiss, M. et al. Contrasting contributions of complementarity-determining region 2 and hypervariable region 4 of rat BV8S2+ (Vβ8.2) TCR to the recognition of myelin basic protein and different types of bacterial superantigens. Int. Immunol. 16, 655–663 (2004).
Hayakawa, K. et al. Positive selection of natural autoreactive B cells. Science 285, 113–116 (1999).
Meyer, S. et al. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell 166, 582–595 (2016).
Mansour, S. et al. Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells. Proc. Natl. Acad. Sci. USA 113, E1266–E1275 (2016).
Zennou, V., Perez-Caballero, D., Göttlinger, H. & Bieniasz, P. D. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78, 12058–12061 (2004).
Fouchier, R. A., Meyer, B. E., Simon, J. H., Fischer, U. & Malim, M. H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16, 4531–4539 (1997).
O’Shea, E. K., Lumb, K. J. & Kim, P. S. Peptide ‘Velcro’: design of a heterodimeric coiled coil. Curr. Biol. 3, 658–667 (1993).
Xu, B. et al. Crystal structure of a γδ T-cell receptor specific for the human MHC class I homolog MICA. Proc. Natl. Acad. Sci. USA 108, 2414–2419 (2011).
Acknowledgements
We are grateful to D. Kaiserlian (INSERM U1111, Lyon) for MODE-K cells; P. Pereira (Institut Pasteur) for the hybridoma (F2.67) producing antibody to Vγ7+ TCR; C. Willcox (University of Birmingham), B. Willcox (University of Birmingham) and P. Barral (The Francis Crick Institute) for cell lines; R.P. Di Marco Barros, A. Jandke, A. Lorenc, D. Ushakov and A. Laing for contributions and discussions; E. Theodoridis, the flow cytometry, genomic equipment park, bio-informatics, experimental histopathology, mass spectrometry and proteomics platform, cell services, and biological service units of the Francis Crick Institute, the Peter Gorer Department of Immunobiology and the Guy’s Hospital Biomedical Research Centre (BRC) for outstanding technical support; and the NVIDIA corporation for the donation of a Titan Xp GPU used to run our protein–protein docking algorithm. The work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (CRUK) (FC001003), the UK Medical Research Council (FC001003), and the Wellcome Trust (FC001003); the CRUK King’s Cancer Centre; studentships from the King’s Bioscience Institute and the Guy’s and St. Thomas’ Charity Prize PhD program in Biomedical and Translational Science (D.M.), the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London (to I.Z.), and the Wellcome Trust (108745/Z/15/Z) (R.J.D.); funds from St. Thomas’ Wegener’s Trust and MRC (MR/P021964/1) (S.J.), the Cluster of Excellence ExC 306 ‘Inflammation-at-Interfaces’ (D.W. and D.K.), Cancer Research UK (23562) (S.M.), and the Wellcome Trust (106292/Z/14/Z and 100156/Z/12/Z) (A.C.H.). This manuscript is dedicated to the memory of Dr. Bruno Kyewski, who greatly clarified our insights into T cell tolerance and selection.
Author information
Authors and Affiliations
Contributions
D.M., I.Z., R.A.G.C., R.J.D., N.A.R., S.J. and P.V. designed and undertook experiments; A.C., O.N., O.P., D.W., D.K. and S.M. designed, prepared and provided critical reagents, D.M., I.Z., R.A.G.C., R.J.D., N.A.R., P.M.I., S.J., S.M., P.A.B., P.V. and A.C.H. processed and interpreted data; D.M., I.Z., R.J.D. and N.A.R revised the manuscript; and P.V. and A.C.H. designed the study and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
O.N. and O.P. are employees of GammdaDelta Therapeutics; and O.N., O.P. and A.C.H. are equity holders in GammaDelta Therapeutics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1-7 and Supplementary Tables 1 and 2
Rights and permissions
About this article
Cite this article
Melandri, D., Zlatareva, I., Chaleil, R.A.G. et al. The γδTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat Immunol 19, 1352–1365 (2018). https://doi.org/10.1038/s41590-018-0253-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-018-0253-5
This article is cited by
-
Comprehensive analysis of BTNL9 as a prognostic biomarker correlated with immune infiltrations in thyroid cancer
BMC Medical Genomics (2023)
-
γδ T cells: origin and fate, subsets, diseases and immunotherapy
Signal Transduction and Targeted Therapy (2023)
-
LncRNA CALML3-AS1 modulated by m6A modification induces BTNL9 methylation to drive non-small-cell lung cancer progression
Cancer Gene Therapy (2023)
-
Local γδ T cells: translating promise to practice in cancer immunotherapy
British Journal of Cancer (2023)
-
Modulation of lytic molecules restrain serial killing in γδ T lymphocytes
Nature Communications (2023)