Abstract
The cytokines IL-17A and IL-17F have 50% amino-acid identity and bind the same receptor; however, their functional differences have remained obscure. Here we found that Il17f–/– mice resisted chemically induced colitis, but Il17a–/– mice did not, and that Il17f−/− CD45RBhiCD4+ T cells induced milder colitis in lymphocyte-deficient Rag2–/– mice, accompanied by an increase in intestinal regulatory T cells (Treg cells). Clostridium cluster XIVa in colonic microbiota capable of inducing Treg cells was increased in both Il17f−/− mice and mice given transfer Il17f−/− T cells, due to decreased expression of a group of antimicrobial proteins. There was substantial production of IL-17F, but not of IL-17A, not only by naive T cells but also by various colon-resident cells under physiological conditions. Furthermore, antibody to IL-17F suppressed the development of colitis, but antibody to IL-17A did not. These observations suggest that IL-17F is an effective target for the treatment of colitis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Yao, Z. et al. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity 3, 811–821 (1995).
Gaffen, S. L., Jain, R., Garg, A. V. & Cua, D. J. The IL-23–IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol. 14, 585–600 (2014).
Iwakura, Y., Ishigame, H., Saijo, S. & Nakae, S. Functional specialization of interleukin-17 family members. Immunity 34, 149–162 (2011).
Ishigame, H. et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30, 108–119 (2009).
Puel, A. et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J. Exp. Med. 207, 291–297 (2010).
Cai, Y. et al. Pivotal role of dermal IL-17-producing γδ T cells in skin inflammation. Immunity 35, 596–610 (2011).
Hueber, W. et al. Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci. Transl. Med. 2, 52ra72 (2010).
Martin, D. A. et al. The emerging role of IL-17 in the pathogenesis of psoriasis: preclinical and clinical findings. J. Invest. Dermatol. 133, 17–26 (2013).
Papp, K. A. et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N. Engl. J. Med. 366, 1181–1189 (2012).
Nakae, S. et al. IL-17 production from activated T cells is required for the spontaneous development of destructive arthritis in mice deficient in IL-1 receptor antagonist. Proc. Natl. Acad. Sci. USA 100, 5986–5990 (2003).
Komiyama, Y. et al. IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J. Immunol. 177, 566–573 (2006).
Yang, X. O. et al. Regulation of inflammatory responses by IL-17F. J. Exp. Med. 205, 1063–1075 (2008).
Starnes, T. et al. Cutting edge: IL-17F, a novel cytokine selectively expressed in activated T cells and monocytes, regulates angiogenesis and endothelial cell cytokine production. J. Immunol. 167, 4137–4140 (2001).
Kawaguchi, M. et al. Identification of a novel cytokine, ML-1, and its expression in subjects with asthma. J. Immunol. 167, 4430–4435 (2001).
Kuestner, R. E. et al. Identification of the IL-17 receptor related molecule IL-17RC as the receptor for IL-17F. J. Immunol. 179, 5462–5473 (2007).
Powrie, F. et al. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity 1, 553–562 (1994).
O’Connor, W. Jr. et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat. Immunol. 10, 603–609 (2009).
Lee, J. S. et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 43, 727–738 (2015).
Maxwell, J. R. et al. Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity 43, 739–750 (2015).
Song, X. et al. Growth factor FGF2 cooperates with interleukin-17 to repair intestinal epithelial damage. Immunity 43, 488–501 (2015).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Tang, C. et al. Inhibition of Dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory t cell expansion in the intestine. Cell Host Microbe 18, 183–197 (2015).
Thornton, A. M. et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184, 3433–3441 (2010).
Gallo, R. L. & Hooper, L. V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12, 503–516 (2012).
Macpherson, A. J., Köller, Y. & McCoy, K. D. The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol. 36, 460–470 (2015).
Pabst, O. New concepts in the generation and functions of IgA. Nat. Rev. Immunol. 12, 821–832 (2012).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011).
Lucke, K., Miehlke, S., Jacobs, E. & Schuppler, M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 55, 617–624 (2006).
Scher, J. U. et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013).
Zhu, J., Yamane, H. & Paul, W. E. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28, 445–489 (2010).
Li, C., Ebert, P. J. & Li, Q. J. T cell receptor (TCR) and transforming growth factor β (TGF-β) signaling converge on DNA (cytosine-5)-methyltransferase to control forkhead box protein 3 (foxp3) locus methylation and inducible regulatory T cell differentiation. J. Biol. Chem. 288, 19127–19139 (2013).
Hooper, L. V., Stappenbeck, T. S., Hong, C. V. & Gordon, J. I. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4, 269–273 (2003).
Qu, X. D. & Lehrer, R. I. Secretory phospholipase A2 is the principal bactericide for staphylococci and other gram-positive bacteria in human tears. Infect. Immun. 66, 2791–2797 (1998).
Okai, S. et al. High-affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat. Microbiol. 1, 16103 (2016).
Hueber, W. et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700 (2012).
Hirota, K. et al. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 14, 372–379 (2013).
Nakae, S. et al. Antigen-specific T cell sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity 17, 375–387 (2002).
Arstila, T. et al. Identical T cell clones are located within the mouse gut epithelium and lamina propia and circulate in the thoracic duct lymph. J. Exp. Med. 191, 823–834 (2000).
Cooper, H. S., Murthy, S. N., Shah, R. S. & Sedergran, D. J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Invest. 69, 238–249 (1993).
Obermeier, F. et al. CpG motifs of bacterial DNA exacerbate colitis of dextran sulfate sodium-treated mice. Eur. J. Immunol. 32, 2084–2092 (2002).
Acknowledgements
We thank Y. Shinkai (Kyoto University) for Rag2−/− mice. Supported by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (Y.I.); by CREST (Y.I.); and by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Y.I. and C.T.).
Author information
Authors and Affiliations
Contributions
C.T. performed the majority of the experimental work and wrote the manuscript in collaboration with M.K., T.K. and T.S.; S. Kakuta performed the antibody-treatment experiments; C.T. and K.S. performed the bacterial 16S rRNA analysis; S. Kubo and S.S. contributed to the generation of Il17f−/−Rag2−/− mouse; H.I. and S.N. contributed to the generation of Il17f−/− mice and Il17a−/− mice, respectively; and Y. I. organized and supervised the project and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Over-expansion of Tregs in Il17f−/− mice accounts for the attenuated colitis.
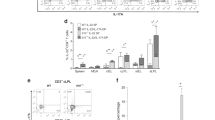
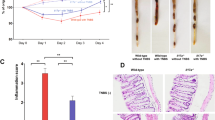
(a, b) Colitis was induced by the method described in Fig. 1c for 42 days, and histological analysis for colonic inflammation (a, H-E staining) and gross appearance (b) were performed (n=3/group). (c) CD25-CD45RBhiCD4+ T cells harvested from BALB/c WT or Il17f−/− mouse drain lymph nodes were transferred into BALB/c Rag2−/− mice for 70 days, and gross appearance were performed (n=4/group). (d-g) The effect of anti-CD25 antibody on the development of DSS-colitis in Il17f–/– mice were examined. (d) The protocol for antibody and DSS treatment. Il17f−/− mice were i.p. administrated with anti-CD25 antibody or control IgG followed by DSS administration. (e) Mice were sacrificed on day 15 after DSS treatment, and colon length and macroscopic histology were examined (n=4/group). *P=0.0476. (f) Gating strategy for the analyzed by flow cytometry. (g) Proportions of total Foxp3+ CD4 T cells in cLP cells, CD25+Foxp3+ and CD25-Foxp3+ Tregs in cLP CD4 T cells were shown (n=5/group). ***P=0.0007, ****P<0.0001. Data in (e, g) are expressed as means ± SD (unpaired two-tailed Student’s t-test).
Supplementary Figure 2 Fecal microbiota compositions are different between Il17f−/− and Il17a−/− mice.
(a) 16S ribosomal RNA sequences of fecal commensal microbiota from WT, Il17f−/− and Il17a−/− mice were analyzed, and proportions of major phyla in the whole microflora were presented (n=9/group). *, #, $: P<0.05. *P=0.0438, **P=0.0054, ***P=0.0009, #P=0.0342, ##P=0.0012, ###P=0.0007, $$P=0.0032.(b, c) Fresh feces from WT, Il17f−/− and Il17a−/− or Il17a−/−Il17f−/− mice were collected and the relative content of total commensal C. XIVa and C. XIVa sp. (b) or commensal Lactobacillus (c) was determined by qPCR (WT n=11, Il17f−/− n=10, Il17a−/− n=4in b; WT n=7, Il17f−/− n=6, Il17a−/−Il17f−/− n=6 in c). *, #: P<0.05.b, *P=0.0435, #P=0.0493, **P=0.0014, ##P=0.0017. c, *P=0.033, #P=0.0106, **P=0.0032. (d) Analysis in (a) were performed, and proportions of indicated species in C. XIVa in the whole microflora are shown. *, #, $: P<0.05. *P=0.028, **P=0.0062, ##P=0.0013, $$P=0.0041. (e) Il17f−/− mice were treated with antibiotics mixture VAMN or vancomycin only for 3 weeks, and fecal microbiota from vancomycin-treated mice were harvested and mixed with C. bolteae (JCM 12243) and C. clostridioforme (JCM 1291) and were transferred into VAMN-treated mice for 3 weeks. Clearance of C. XIVa in vancomycin-treated mouse feces were confirmed, and cLP Treg population in recipient mice after microbiota transfer was examined by flow cytometry (n=3/group). *, #, $, &: P<0.05. *P=0.0476, #P=0.0306, $P=0.0482, &P=0.0455. (f) SPF WT mice were orally administrated with a mixture of C. bolteae (JCM 12243) and C. clostridioforme (JCM 1291) or P. loescheii (JCM 12249) and P. shahii (JCM 12083) every 2 days for 2 weeks, and relative levels of indicated mRNA in the colon were determined by qPCR (n=4/group). *, #, $, &: P<0.05. *P=0.0251, **P=0.0052, #P=0.035, $P=0.0154, &P=0.0211. (g) Analysis in (a) were performed, and proportions of a part of C. IV species in the whole microflora are shown (n=9/group). Data in (b, e, f) are expressed as means ± SD, and measure of center in (a, c, d, g) is mean (for a-f, one-way ANOVA followed by Tukey’s multiple-comparisons test).
Supplementary Figure 3 IL-17F regulates the colonization of C. XIVa.
(a) Three week-old WT and Il17f–/– mice were weaned and co-housed in the same cages for 4 weeks. Then, they were separated into different cages. After the indicated period, fecal microbiota was harvested and the relative content of C. XIVa and Prevotella was determined by qPCR (WT n=7, Il17f−/− n=5). *P=0.0119, **P=0.0057 (unpaired two-tailed Student’s t-test). (b) The expression of indicated genes encoding AMPs in WT, Il17f−/− or Il17a−/− mouse colon was examined by qPCR (in panels from Lcn2 to Defa4, n=4/group; in other panels, WT n=12, Il17f–/– n=8, Il17a–/– n=8). *, #: P<0.05. *P=0.0333, **P=0.007, ***P=0.0005, #P=0.0433 (one-way ANOVA followed by Tukey’s multiple-comparisons test). Data are expressed as means ± SD.
Supplementary Figure 4 Dose dependency of the effect of AMPs on bacterial growth.
(a) C. bolteae, C. clostridioforme, P. loescheii or P. shahii was cultured in the present of indicated AMPs with different concentrations for 48 hr, and bacterial growth/recovery was measured with spectrophotometer (3 replicates for each sample). *, #, $, &: P<0.05. In +Angionenin4 panels, **P=0.0068, ***P=0.0001, ##P=0.0096, $$P=0.0056; in +PhospholipaseA2 panels, **P=0.0049, ***P=0.001, ##P=0.0038, $$P=0.0026; in +β-denfensin1 panels, **P=0.0037, ***P=0.0001, ##P=0.0017, ###P=0.0006, $$$P=0.0002, &&&P=0.0001; in +β-denfensin4 panels, *P=0.0262, **P=0.0011, ***P=0.0002, #P=0.0142, ##P=0.0017, ###P=0.0007. (b-d) C. bolteae, C. clostridioforme, P. loescheii, P. shahii, Lactobacillus murinus or Alcaligenes faecalis was cultured in the presence of indicated AMPs (5 mg/ml of each AMP), and bacterial growth/recovery at indicated time points was measured with spectrophotometer (3 replicates for each sample). b, *P=0.0275. c, *P=0.0123, **P=0.0021. Data are expressed as means ± SD (one-way ANOVA followed by Tukey’s multiple-comparisons test).
Supplementary Figure 5 Intestinal IgA production is normal in Il17f−/− mice.
(a, b) Proportion of IgA+ B cells in the colon of indicated mice were examined by flow cytometry (n=3–6/group). (a) Cells are pre-gated on cLP 7AAD-Lymphocytes (left) or 7AAD-CD19+Lymphocytes (right) and numbers in dot plot panels indicate percentages of IgA+ B cells in total cLPLs (left) or total cLP B cells (right). (b) Percentages of IgA+ B cells in cLP total B cells in indicated mice are shown (WT n=4, l17f–/– n=3, Il17a–/– n=5, l17f–/–Il17a–/– n=3). WT vs Il17a–/– *P=0.0158, Il17f–/– vs Il17a–/–Il17f–/– *P=0.0356. (c) Expression of Pigr in the colon of indicated mice were examined by qPCR (WT n=12, l17f–/– n=8, Il17a–/– 7). ***P=0.0006. (d-f) Fecal microflora were harvested from WT, Il17f−/− or Il17a−/− mice and were stained by anti-IgA Ab. The IgA+ or IgA- commensal bacteria were purified by FACS sorting, and were further identified as IgA-specific/nonspecific commensals. (d) IgA-bound/non-bound bacterial population before and after the purification was confirmed by flow cytometry. (e) Indicated IgA-specific/nonspecific commensals in indicated groups of mice are shown by the ratio of % in purified total IgA+ bacteria / % in purified total IgA- bacteria. (f) IgA-specific/nonspecific commensals in indicated individual mouse are shown in heat map by using the same data described in (e) (n=3/group). Data in (a, d) are representative of two independent experiments with similar results. Data in (b, c, e) are expressed as means ± SD (for b-c, one-way ANOVA followed by Tukey’s multiple-comparisons test).
Supplementary Figure 6 C. XIVa can suppress commensal Prevotella growth.
(a) The content of Prevotellaceae species the whole microflora is shown (n=9/group). **P=0.0006. Measure of center in each group is mean. (b) Experiment described in Fig. 5e was carried out. Mice were transferred with a mixture of C. bolteae and C. clostridioforme and the proportion of fecal Actinobacteria, Proteobacteria and Lactobacillus was examined by qPCR (n=4/group). (c) Colon pieces from WT mice were stimulated with a mixture of C. XIV (equal CFU of C. clostridioforme + C. bolteae) with indicated doses for 11 hr, and the expression of indicated AMPs was determined by qPCR (3 replicates for each sample). Data in (c) are expressed as means ± SD (for a&c, one-way ANOVA followed by Tukey’s multiple-comparisons test).
Supplementary Figure 7 Correlation between Il17a and Il17f expression and their associated transcription factors.
(a) Lymphocytes isolated from WT or Il17a−/−Il17f−/− mouse spleens were stimulated with or without PMA + Ionomycin for 5 h, and IL-17F- or IL-17A-producing CD4+ T cell subsets were determined by flow cytometry. Data in are representative of two independent experiments with similar results. (b) CD25-CD45RBhi CD4 T cells were sorted from WT or Il17f−/− mouse spleen and the mRNA expression of Il17f, Il17a, Il17ra and Il17rc was examined by real-time qPCR (n=4/group). (c) CD62L+CD4+ naïve T cells purified from WT spleen and LNs were stimulated with anti-CD3 and anti-CD28 Abs, and Il17a and Il17f expression were determined at indicated time points by qPCR. Relative values to naïve T cells are shown (2 replicates for each sample). (d, e) Messenger RNA levels of Il17a and Il17f and their associated transcription factors, Irf4, IκBζ, RoRγt, Runx1 and Batf, in colonic cells from lamina propria or intraepithelial layer described in Fig. 7c were examined by real-time qPCR, and the correlations between Il17a or Il17f and each transcription factor are shown (11 samples in each panel). In the right column of (e), correlation was calculated using colonic cells excluding cLP NK and DCs (8 samples in each panel). Data in (b, c) are expressed as means ± SD.
Supplementary information
Supplementary Figures and Text
Supplementary Figures 1-7 and Supplementary Tables 1 and 2
Rights and permissions
About this article
Cite this article
Tang, C., Kakuta, S., Shimizu, K. et al. Suppression of IL-17F, but not of IL-17A, provides protection against colitis by inducing Treg cells through modification of the intestinal microbiota. Nat Immunol 19, 755–765 (2018). https://doi.org/10.1038/s41590-018-0134-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-018-0134-y
This article is cited by
-
Strategies for targeting cytokines in inflammatory bowel disease
Nature Reviews Immunology (2024)
-
IL-17 and IL-17-producing cells in protection versus pathology
Nature Reviews Immunology (2023)
-
Blocking Dectin-1 prevents colorectal tumorigenesis by suppressing prostaglandin E2 production in myeloid-derived suppressor cells and enhancing IL-22 binding protein expression
Nature Communications (2023)
-
The IL-17 family in diseases: from bench to bedside
Signal Transduction and Targeted Therapy (2023)
-
Nsun2 coupling with RoRγt shapes the fate of Th17 cells and promotes colitis
Nature Communications (2023)