Abstract
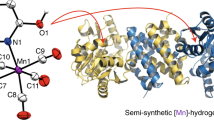
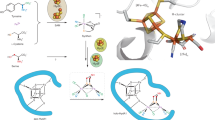
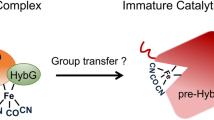
[NiFe]-hydrogenases are biotechnologically relevant enzymes catalyzing the reversible splitting of H2 into 2e− and 2H+ under ambient conditions. Catalysis takes place at the heterobimetallic NiFe(CN)2(CO) center, whose multistep biosynthesis involves careful handling of two transition metals as well as potentially harmful CO and CN− molecules. Here, we investigated the sequential assembly of the [NiFe] cofactor, previously based on primarily indirect evidence, using four different purified maturation intermediates of the catalytic subunit, HoxG, of the O2-tolerant membrane-bound hydrogenase from Cupriavidus necator. These included the cofactor-free apo-HoxG, a nickel-free version carrying only the Fe(CN)2(CO) fragment, a precursor that contained all cofactor components but remained redox inactive and the fully mature HoxG. Through biochemical analyses combined with comprehensive spectroscopic investigation using infrared, electronic paramagnetic resonance, Mössbauer, X-ray absorption and nuclear resonance vibrational spectroscopies, we obtained detailed insight into the sophisticated maturation process of [NiFe]-hydrogenase.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the article and the Supplementary Information. Raw XAS data were obtained at the SSRL synchrotron. Raw NRVS data were generated at the synchrotron facilities Petra III and SPring-8, and are available in their processed form upon request. Additional reagents will be made available by the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Lubitz, W., Ogata, H., Rüdiger, O. & Reijerse, E. Hydrogenases. Chem. Rev. 114, 4081–4148 (2014).
Peters, J. W. et al. [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim. Biophys. Acta 1853, 1350–1369 (2015).
Vignais, P. M. & Billoud, B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272 (2007).
Britt, R. D., Rao, G. & Tao, L. Biosynthesis of the catalytic H-cluster of [FeFe] hydrogenase: the roles of the Fe–S maturase proteins HydE, HydF, and HydG. Chem. Sci. 11, 10313–10323 (2020).
Shima, S., Fujishiro, T. & Ermler, U. in Biohydrogen (ed. Rögner, M.) 127–144 (de Gruyter, 2015).
Böck, A., King, P. W., Blokesch, M. & Posewitz, M. C. in Advances in Microbial Physiology Vol. 51 (ed. Poole, R. K.) Ch. 1 (Elsevier, 2006).
Lacasse, M. J. & Zamble, D. B. [NiFe]-Hydrogenase maturation. Biochemistry 55, 1689–1701 (2016).
Britt, R. D., Rao, G. & Tao, L. Bioassembly of complex iron–sulfur enzymes: hydrogenases and nitrogenases. Nat. Rev. Chem. 4, 542–549 (2020).
Fritsch, J., Lenz, O. & Friedrich, B. Structure, function and biosynthesis of O2-tolerant hydrogenases. Nat. Rev. Microbiol. 11, 106–114 (2013).
Caserta, G. et al. Unusual structures and unknown roles of FeS clusters in metalloenzymes seen from a resonance Raman spectroscopic perspective. Coord. Chem. Rev. 452, 214287 (2022).
Marques, M. C. et al. The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis. Nat. Chem. Biol. 13, 544–550 (2017).
Pierik, A. J., Roseboom, W., Happe, R. P., Bagley, K. A. & Albracht, S. P. J. Carbon monoxide and cyanide as intrinsic ligands to iron in the active site of [NiFe]-hydrogenases. J. Biol. Chem. 274, 3331–3337 (1999).
Frielingsdorf, S. et al. Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase. Nat. Chem. Biol. 10, 378–385 (2014).
Ogata, H., Nishikawa, K. & Lubitz, W. Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase. Nature 520, 571–574 (2015).
Miki, K., Atomi, H. & Watanabe, S. Structural insight into [NiFe] hydrogenase maturation by transient complexes between Hyp proteins. Acc. Chem. Res. 53, 875–886 (2020).
Bürstel, I. et al. A universal scaffold for synthesis of the Fe(CN)2(CO) moiety of [NiFe] hydrogenase. J. Biol. Chem. 287, 38845–38853 (2012).
Soboh, B. et al. [NiFe]-hydrogenase maturation: isolation of a HypC-HypD complex carrying diatomic CO and CN− ligands. FEBS Lett. 586, 3882–3887 (2012).
Reissmann, S. Taming of a poison: biosynthesis of the NiFe-hydrogenase cyanide ligands. Science 299, 1067–1070 (2003).
Schulz, A.-C. et al. Formyltetrahydrofolate decarbonylase synthesizes the active site CO ligand of O2-tolerant [NiFe] hydrogenase. J. Am. Chem. Soc. 142, 1457–1464 (2020).
Bürstel, I. et al. CO synthesized from the central one-carbon pool as source for the iron carbonyl in O2-tolerant [NiFe]-hydrogenase. Proc. Natl Acad. Sci. USA 113, 14722–14726 (2016).
Senger, M., Stripp, S. T. & Soboh, B. Proteolytic cleavage orchestrates cofactor insertion and protein assembly in [NiFe]-hydrogenase biosynthesis. J. Biol. Chem. 292, 11670–11681 (2017).
Stripp, S. T. et al. Electron inventory of the iron–sulfur scaffold complex HypCD essential in [NiFe]-hydrogenase cofactor assembly. Biochem. J. 478, 3281–3295 (2021).
Arlt, C. et al. Native mass spectrometry identifies the HybG chaperone as carrier of the Fe(CN)2CO group during maturation of E. coli [NiFe]-hydrogenase 2. Sci. Rep. 11, 24362 (2021).
Watanabe, S. et al. Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer. Proc. Natl Acad. Sci. USA 112, 7701–7706 (2015).
Hartmann, S., Frielingsdorf, S., Caserta, G. & Lenz, O. A membrane-bound [NiFe]-hydrogenase large subunit precursor whose C-terminal extension is not essential for cofactor incorporation but guarantees optimal maturation. MicrobiologyOpen 9, 1197–1206 (2020).
Pinske, C., Thomas, C., Nutschan, K. & Sawers, R. G. Delimiting the function of the C-terminal extension of the Escherichia coli [NiFe]-hydrogenase 2 large subunit precursor. Front. Microbiol. 10, 2223 (2019).
Pinske, C. & Sawers, R. G. Anaerobic formate and hydrogen metabolism. EcoSal, https://doi.org/10.1128/ecosalplus.ESP-0011-2016 (2016).
Blokesch, M. & Böck, A. Maturation of [NiFe]-hydrogenases in Escherichia coli: the HypC cycle. J. Mol. Biol. 324, 287–296 (2002).
Fritsch, J. et al. The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre. Nature 479, 249–252 (2011).
Lenz, O., Lauterbach, L., Frielingsdorf, S. & Friedrich, B. in Biohydrogen (ed. Rögner, M.) 61–88 (de Gruyter, 2015).
Vincent, K. A. et al. Electrocatalytic hydrogen oxidation by an enzyme at high carbon monoxide or oxygen levels. Proc. Natl Acad. Sci. USA 102, 16951–16954 (2005).
Hartmann, S. et al. O2-tolerant H2 activation by an isolated large subunit of a [NiFe] hydrogenase. Biochemistry 57, 5339–5349 (2018).
Caserta, G. et al. The large subunit of the regulatory [NiFe]-hydrogenase from Ralstonia eutropha – a minimal hydrogenase? Chem. Sci. 11, 5453–5465 (2020).
Eberz, G., Eitinger, T. & Friedrich, B. Genetic determinants of a nickel-specific transport system are part of the plasmid-encoded hydrogenase gene cluster in Alcaligenes eutrophus. J. Bacteriol. 171, 1340–1345 (1989).
Bernhard, M., Schwartz, E., Rietdorf, J. & Friedrich, B. The Alcaligenes eutrophus membrane-bound hydrogenase gene locus encodes functions involved in maturation and electron transport coupling. J. Bacteriol. 178, 4522–4529 (1996).
Rossmann, R., Sauter, M., Lottspeich, F. & Böck, A. Maturation of the large subunit (HYCE) of Escherichia coli hydrogenase 3 requires nickel incorporation followed by C-terminal processing at Arg537. Eur. J. Biochem. 220, 377–384 (1994).
Kwon, S. et al. Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA. Proc. Natl Acad. Sci. USA 115, 7045–7050 (2018).
Roncaroli, F. et al. Cofactor composition and function of a H2-sensing regulatory hydrogenase as revealed by Mössbauer and EPR spectroscopy. Chem. Sci. 6, 4495–4507 (2015).
Caserta, G. et al. Hydroxy-bridged resting states of a [NiFe]-hydrogenase unraveled by cryogenic vibrational spectroscopy and DFT computations. Chem. Sci. 12, 2189–2197 (2021).
Caserta, G. et al. In vitro assembly as a tool to investigate catalytic intermediates of [NiFe]-hydrogenase. ACS Catal. 10, 13890–13894 (2020).
Ogata, H. et al. Hydride bridge in [NiFe]-hydrogenase observed by nuclear resonance vibrational spectroscopy. Nat. Commun. 6, 7890 (2015).
Lorent, C. et al. Exploring structure and function of redox intermediates in [NiFe]‐hydrogenases by an advanced experimental approach for solvated, lyophilized and crystallized metalloenzymes. Angew. Chem. Int. Ed. Engl. 60, 15854–15862 (2021).
Krämer, T., Kampa, M., Lubitz, W., van Gastel, M. & Neese, F. Theoretical spectroscopy of the NiII intermediate states in the catalytic cycle and the activation of [NiFe] hydrogenases. ChemBioChem 14, 1898–1905 (2013).
Blokesch, M. & Böck, A. Properties of the [NiFe]-hydrogenase maturation protein HypD. FEBS Lett. 580, 4065–4068 (2006).
Albareda, M., Pacios, L. F. & Palacios, J. M. Computational analyses, molecular dynamics, and mutagenesis studies of unprocessed form of [NiFe] hydrogenase reveal the role of disorder for efficient enzyme maturation. Biochim. Biophys. Acta Bioenerg. 1860, 325–340 (2019).
Magalon, A. & Böck, A. Analysis of the HypC-HycE complex, a key intermediate in the assembly of the metal center of the Escherichia coli hydrogenase 3. J. Biol. Chem. 275, 21114–21120 (2000).
Massanz, C. & Friedrich, B. Amino acid replacements at the H2-activating site of the NAD-reducing hydrogenase from Alcaligenes eutrophus. Biochemistry 38, 14330–14337 (1999).
Winter, G., Buhrke, T., Jones, A. K. & Friedrich, B. The role of the active site-coordinating cysteine residues in the maturation of the H2-sensing [NiFe] hydrogenase from Ralstonia eutropha H16. Arch. Microbiol. 182, 138–46 (2004).
Esselborn, J. et al. Spontaneous activation of [FeFe]-hydrogenases by an inorganic [2Fe] active site mimic. Nat. Chem. Biol. 9, 607–609 (2013).
Shima, S. et al. Reconstitution of [Fe]-hydrogenase using model complexes. Nat. Chem. 7, 995–1002 (2015).
Lenz, O. & Friedrich, B. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl Acad. Sci. USA 95, 12474–12479 (1998).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Lenz, O., Lauterbach, L. & Frielingsdorf, S. in Methods in Enzymology Vol. 613 (ed. Armstrong, F) Ch. 5 (Elsevier, 2018).
Fritsch, J., Lenz, O. & Friedrich, B. The maturation factors HoxR and HoxT contribute to oxygen tolerance of membrane-bound [NiFe] hydrogenase in Ralstonia eutropha H16. J. Bacteriol. 193, 2487–2497 (2011).
Kleihues, L., Lenz, O., Bernhard, M., Buhrke, T. & Friedrich, B. The H2 sensor of Ralstonia eutropha is a member of the subclass of regulatory [NiFe] hydrogenases. J. Bacteriol. 182, 2716–2724 (2000).
Wolf, I., Buhrke, T., Dernedde, J., Pohlmann, A. & Friedrich, B. Duplication of hyp genes involved in maturation of [NiFe] hydrogenases in Alcaligenes eutrophus H16. Arch. Microbiol 170, 451–459 (1998).
Dernedde, J., Eitinger, T., Patenge, N. & Friedrich, B. hyp gene products in Alcaligenes eutrophus are part of a hydrogenase-maturation system. Eur. J. Biochem. 235, 351–358 (1996).
Buhrke, T. Der H2-Sensor von Ralstonia eutropha: Struktur-Funktionsbeziehungen einer neuartigen [NiFe]-Hydrogenase. PhD Thesis, Humboldt-Universität zu Berlin (2002).
Bernhard, M. Maturationswege oligomerer kofaktorhaltiger Redoxenzyme am Beispiel der drei [NiFe] Hydrogenasen von Ralstonia eutropha. PhD thesis, Humboldt-Universität zu Berlin (2000).
Stoll, S. & Schweiger, A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 178, 42–55 (2006).
Gee, L. B., Wang, H. & Cramer, S. P. in Methods in Enzymology Vol. 599 (ed. David, S.) 409–425 (Elsevier, 2018).
Acknowledgements
G.C., S.H., O.L., I.Z. and P.H. are grateful to the Einstein Foundation Berlin (grant number EVF-2016-277) for funding. This work was also supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through the PP 1927 ‘Iron Sulfur for Life’ (project no. DE1877/1-1 (S.D.), 311062227 (O.L., I.Z.)) and the cluster of excellence ‘UniSysCat’ under Germany’s Excellence Strategy-EXC2008-390540038. The authors are indebted for EU financial support (Article 38.1.2, GA) within the European Union’s Horizon 2020 research and innovation program under grant agreement no. 810856. C.v.S. and S.D. thank the Max-Planck Society for funding. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. C.v.S. and S.D. gratefully acknowledge M. Latimer for his support and assistance during XAS measurements at beamline 9-3. NRVS data collection was supported by the proposal at BL09XU [2019A1201] at SPring-8. We acknowledge DESY (Hamburg, Germany), a member of the Helmholtz Association HGF, for the provision of experimental facilities. Parts of this research were carried out at Petra III and we thank A. Jafari for assistance in using P01. Beamtime was allocated for proposal I-20200452. J.-P. Oudsen is gratefully acknowledged for assistance in the NRVS data acquisition. We thank S. P. Cramer for fruitful discussions on NRVS data and S. Leimkühler for ICP-OES measurements. We are grateful to J. Fritsch for performing preliminary experiments on the HoxG intermediates, and to A. C. Schulz for HypCD sample preparations.
Author information
Authors and Affiliations
Contributions
G.C., S.H., S.F. and O.L. conceived and designed experiments. G.C., S.H., S.F. and J.S. conducted molecular biology experiments. S.H. and G.C. performed sample preparations and biochemical assays. G.C. performed sample preparation for synchrotron measurements. G.C., S.H., C.K.-R. and I.Z. performed and analyzed IR spectroscopic experiments. C. Lorent performed and analyzed EPR measurements. M.K., S.Y. and C. Limberg performed and analyzed Mössbauer experiments. G.C., Y.Y. and I.S. acquired and analyzed NRVS data. C.v.S. and S.D. performed and analyzed EXAFS experiments. G.C., S.H., O.L, S.F., I.Z. and P.H. analyzed the data. G.C., S.H., S.F. and O.L. wrote the manuscript with input from all co-authors. All authors have given approval to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–6, Figs. 1–14, note and refs.
Supplementary Data 1
Contains source data of Supplementary Fig. 2.
Supplementary Data 2
Contains source data of Supplementary Fig. 3.
Supplementary Data 3
Contains source data of Supplementary Fig. 4.
Supplementary Data 4
Contains source data of Supplementary Fig. 10.
Supplementary Data 5
Contains source data of Supplementary Fig. 11.
Supplementary Data 6
Contains source data of Supplementary Fig. 12.
Supplementary Data 7
Contains source data of Supplementary Fig. 13.
Source data
Source Data Fig. 2
Contains source data of Fig. 2c
Source Data Fig. 3
Contains source data of Fig. 3.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Caserta, G., Hartmann, S., Van Stappen, C. et al. Stepwise assembly of the active site of [NiFe]-hydrogenase. Nat Chem Biol 19, 498–506 (2023). https://doi.org/10.1038/s41589-022-01226-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01226-w