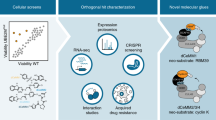
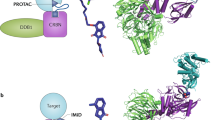
Abstract
Targeted protein degradation is a novel pharmacology established by drugs that recruit target proteins to E3 ubiquitin ligases. Based on the structure of the degrader and the target, different E3 interfaces are critically involved, thus forming defined ‘functional hotspots’. Understanding disruptive mutations in functional hotspots informs on the architecture of the assembly, and highlights residues susceptible to acquire resistance phenotypes. Here we employ haploid genetics to show that hotspot mutations cluster in substrate receptors of hijacked ligases, where mutation type and frequency correlate with gene essentiality. Intersection with deep mutational scanning revealed hotspots that are conserved or specific for chemically distinct degraders and targets. Biophysical and structural validation suggests that hotspot mutations frequently converge on altered ternary complex assembly. Moreover, we validated hotspots mutated in patients that relapse from degrader treatment. In sum, we present a fast and widely accessible methodology to characterize small-molecule degraders and associated resistance mechanisms.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Raw and analysed mutational scanning and hybrid-capture datasets (Figs. 1–5 and Supplementary Figs 1,3–5) are available in the Gene Expression Omnibus database under accession code GSE198280. For their analysis the human reference genome (hg38/GRCh38 assembly, GenBank accession 883148) was used. Atomic coordinates and structure factors for the new protein structure VCB–AT7–Brd4BD2 is available at the PDB under accession 7ZNT. All data generated and analysed in this study are included in this published article, its Supplementary Information, the mentioned databases or are available from the corresponding authors upon request. Source data are provided with this paper.
Code availability
All code used for analysis of the experimental data is available at https://github.com/GWinterLab/TPDR.
References
Deshaies, R. J. Multispecific drugs herald a new era of biopharmaceutical innovation. Nature 580, 329–338 (2020).
Gerry, C. J. & Schreiber, S. L. Unifying principles of bifunctional, proximity-inducing small molecules. Nat. Chem. Biol. 16, 369–378 (2020).
Hanzl, A. & Winter, G. E. Targeted protein degradation: current and future challenges. Curr. Opin. Chem. Biol. 56, 35–41 (2020).
Petroski, M. D. & Deshaies, R. J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005).
Harper, J. W. & Schulman, B. A. Cullin-RING ubiquitin ligase regulatory circuits: a quarter century beyond the F-box hypothesis. Annu. Rev. Biochem. 90, 403–429 (2021).
Kramer, L. T. & Zhang, X. Expanding the landscape of E3 ligases for targeted protein degradation. Curr. Res. Chem. Biol. 2, 100020 (2022).
Békés, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Kozicka, Z. & Thomä, N. H. Haven’t got a glue: Protein surface variation for the design of molecular glue degraders. Cell Chem. Biol. 28, 1032–1047 (2021).
Maniaci, C. & Ciulli, A. Bifunctional chemical probes inducing protein-protein interactions. Curr. Opin. Chem. Biol. 52, 145–156 (2019).
Gadd, M. S. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017).
Nowak, R. P. et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 14, 706–714 (2018).
Zorba, A. et al. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc. Natl Acad. Sci. USA 115, E7285–E7292 (2018).
Shirasaki, R. et al. Functional genomics identify distinct and overlapping genes mediating resistance to different classes of heterobifunctional degraders of oncoproteins. Cell Rep. 34, 108532 (2021).
Sievers, Q. L., Gasser, J. A., Cowley, G. S., Fischer, E. S. & Ebert, B. L. Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4(CRBN) activity. Blood 132, 1293–1303 (2018).
Mayor-Ruiz, C. et al. Plasticity of the Cullin-RING ligase repertoire shapes sensitivity to ligand-induced protein degradation. Mol. Cell 75, 849–858 (2019).
Zhang, L., Riley-Gillis, B., Vijay, P. & Shen, Y. Acquired resistance to BET-PROTACs (proteolysis-targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol. Cancer Ther. 18, 1302–1311 (2019).
Ferguson, F. M. & Gray, N. S. Kinase inhibitors: the road ahead. Nat. Rev. Drug Discov. 17, 353–377 (2018).
Zaidman, D., Prilusky, J. & London, N. ProsetTac: Rosetta based modeling of PROTAC mediated ternary complexes. J. Chem. Inf. Model. 60, 4894–4903 (2020).
Bai, N. et al. Rationalizing PROTAC-mediated ternary complex formation using Rosetta. J. Chem. Inf. Model. 61, 1368–1382 (2021).
Drummond, M. L. & Williams, C. I. In silico modeling of PROTAC-mediated ternary complexes: validation and application. J. Chem. Inf. Model. 59, 1634–1644 (2019).
Sievers, Q. L. et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362, 2018 (1979).
Eron, S. J. et al. Structural characterization of degrader-induced ternary complexes using hydrogen-deuterium exchange mass spectrometry and computational modeling: implications for structure-based design. ACS Chem. Biol. 16, 2228–2243 (2021).
Dixon, T. et al. Predicting the structural basis of targeted protein degradation by integrating molecular dynamics simulations with structural mass spectrometry. Nat. Commun. 13, 5884 (2022).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
Latif, F. et al. Identification of the von Hippel–Lindau disease tumor suppressor gene. Science 260, 1317–1320 (1993).
Raina, K. et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl Acad. Sci. USA 113, 7124–7129 (2016).
Winter, G. E. et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol. Cell 67, 5–18 (2017).
Forment, J. V. et al. Genome-wide genetic screening with chemically mutagenized haploid embryonic stem cells. Nat. Chem. Biol. 13, 12–14 (2017).
Volz, J. C., Schuller, N. & Elling, U. Using functional genetics in haploid cells for drug target identification. Methods Mol. Biol. 1953, 3–21 (2019).
Winter, G. E. et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat. Chem. Biol. 10, 768–773 (2014).
Suiter, C. C. et al. Massively parallel variant characterization identifies NUDT15 alleles associated with thiopurine toxicity. Proc. Natl Acad. Sci. USA 117, 5394–5401 (2020).
Awad, M. M. et al. Acquired resistance to KRAS G12C inhibition in cancer. N. Engl. J. Med. 384, 2382–2393 (2021).
Zengerle, M., Chan, K. H. & Ciulli, A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 10, 1770–1777 (2015).
Testa, A., Hughes, S. J., Lucas, X., Wright, J. E. & Ciulli, A. Structure-based design of a macrocyclic PROTAC. Angew. Chem. Int. Ed. Engl. 59, 1727–1734 (2020).
Farnaby, W. et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 15, 672–680 (2019).
Soares, P. et al. Group-based optimization of potent and cell-active inhibitors of the von Hippel–Lindau (VHL) E3 ubiquitin ligase: structure-activity relationships leading to the chemical probe (2S,4R)-1-((S)-2-(1-cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298). J. Med. Chem. 61, 599–618 (2018).
Matyskiela, M. E. et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature 535, 252–257 (2016).
Surka, C. et al. CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells. Blood 137, 661–677 (2021).
Fink, E. C. et al. Crbn I391V is sufficient to confer in vivo sensitivity to thalidomide and its derivatives in mice. Blood 132, 1535–1544 (2018).
Olson, C. M. et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat. Chem. Biol. 14, 163–170 (2017).
Barrio, S. et al. IKZF1/3 and CRL4 CRBN E3 ubiquitin ligase mutations and resistance to immunomodulatory drugs in multiple myeloma. Haematologica 105, E237–E241 (2020).
Gooding, S. et al. Multiple cereblon genetic changes are associated with acquired resistance to lenalidomide or pomalidomide in multiple myeloma. Blood 137, 232–237 (2021).
Roy, M. J. et al. SPR-measured dissociation kinetics of PROTAC ternary complexes influence target degradation rate. ACS Chem. Biol. 14, 361–368 (2019).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Scholes, N. S., Mayor-Ruiz, C. & Winter, G. E. Identification and selectivity profiling of small-molecule degraders via multi-omics approaches. Cell Chem. Biol. 28, 1048–1060 (2021).
Klein, V. G., Bond, A. G., Craigon, C., Lokey, R. S. & Ciulli, A. Amide-to-ester substitution as a strategy for optimizing PROTAC permeability and cellular activity. J. Med. Chem. 64, 18082–18101 (2021).
Jiang, B. et al. Discovery and resistance mechanism of a selective CDK12 degrader. Nat. Chem. Biol. 17, 675–683 (2021).
Gosavi, P. M. et al. Profiling the landscape of drug resistance mutations in neosubstrates to molecular glue degraders. ACS Cent. Sci. 8, 417–429 (2022).
Kortum, K. M. et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood 128, 1226–1233 (2016).
Galdeano, C. et al. Structure-guided design and optimization of small molecules targeting the protein-protein interaction between the von Hippel–Lindau (VHL) E3 ubiquitin ligase and the hypoxia inducible factor (HIF) α subunit with in vitro nanomolar affinities. J. Med. Chem. 57, 8657–8663 (2014).
Joung, J. et al. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Guzmán, C., Bagga, M., Kaur, A., Westermarck, J. & Abankwa, D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS ONE 9, e92444 (2014).
Barnett, D. W., Garrison, E. K., Quinlan, A. R., Str̈mberg, M. P. & Marth, G. T. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27, 1691–1692 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McLaren, W. et al. The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
van Molle, I. et al. Dissecting fragment-based lead discovery at the von Hippel–Lindau protein: hypoxia inducible factor 1α protein–protein interface. Chem. Biol. 19, 1300–1312 (2012).
Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 (2003).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
We thank the Biomedical Sequencing Facility at CeMM for assistance with next-generation sequencing, C. Crowe (Ciulli laboratory) for the gift of purified BET-bromodomain protein, the Diamond Light Source for beamtime (BAG proposal MX14980-13) and J. Bigenzahn (Superti-Furga laboratory) for the gift of plasmids. CeMM and the Winter laboratory are supported by the Austrian Academy of Sciences. The Winter laboratory is further supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement 851478), as well as by funding from the Austrian Science Fund (FWF, projects P32125, P31690 and P7909). The work of the Ciulli laboratory on PROTACs has received funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013) as a Starting Grant to A.C. (grant agreement ERC-2012-StG-311460 DrugE3CRLs). R.C. is funded by a PhD studentship from the UK Biotechnology and Biological Sciences Research Council (BBSRC) under the EastBio doctoral training programme (BB/M010996/1). Biophysics and drug-discovery activities at Dundee were supported by Wellcome Trust strategic awards 100476/Z/12/Z and 094090/Z/10/Z, respectively. For the purpose of open access, the authors have applied a Creative Commons Attribution (CC-BY) licence to any author-accepted manuscript version arising.
Author information
Authors and Affiliations
Contributions
A.H., M.B. and G.E.W. conceptualized this study. A.H. and M.B. designed and conducted hybrid-capture assays. A.H., S.B. and M.B. designed and conducted deep mutational scanning assays. A.H., S.B. and E.B. generated cell lines and conducted cellular mutant validation including immunoblotting and drug sensitivity assays. M.B. and H.I. analyzed and visualized hybrid capture and deep mutational scanning data. A.C. and A.T. designed AT7 compound and A.T. synthesized the compound. R.C. expressed and purified recombinant proteins, performed fluorescence polarization measurements and compound synthesis. S.J.H. solved co-crystal structure. J.W. performed degradation and cell viability assays for AT7. A.C. and G.E.W. supervised the work. H.I., A.H. and R.C. generated figures with input from all authors. A.H., R.C., A.C. and G.E.W. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
S.B. is an employee at Proxygen, a company that is developing molecular glue degraders. M.B. is scientific founder, shareholder and employee at Proxygen. G.E.W. is scientific founder and shareholder at Proxygen and Solgate and the Winter laboratory receives research funding from Pfizer. A.C. is a scientific founder, shareholder and advisor of Amphista Therapeutics, a company that is developing targeted protein degradation therapeutic platforms. S.J.H. and A.T. are currently employees of Amphista Therapeutics. The Ciulli laboratory receives or has received sponsored research support from Almirall, Amgen, Amphista Therapeutics, Boehringer Ingelheim, Eisai, Merck KaaG, Nurix Therapeutics, Ono Pharmaceutical and Tocris-Biotechne. The other authors are not aware of any affiliations, memberships, funding or financial holdings that might be perceived as affecting the objectivity of this work.
Peer review
Peer review information
Nature Chemical Biology thanks Frank Sicheri, Jun Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Quantitative and Qualitative Differences in Degrader Resistance.
(a) Dose-resolved, normalized viability after 3 d treatment (dBET6 or ARV-771) in KBM7, MV4;11 and MOLM-13 cells. Mean ± s.e.m.; n = 3 independent treatments. (b) Histogram depicting growth competition experiments. WT control KBM7 cells were mixed with mCherry and Cas9 expressing KBM7 cells harboring sgRNAs against the indicated genes. Pools were flow cytometry quantified at days 0, 7, 14 and 21 and mCherry percentages were normalized to day 0 percentage and to a non-targeting control sgRNA (sgMYCdesert). Data represents mean ± s.d. of n = 3 biological replicates. (c) Scheme of targeted hybrid-capture approach coupled to next-generation sequencing to identify mutations in spontaneously resistant cells. (d) Structure depiction of the CUL2-VBC-MZ1-BRD4 complex (PDBs: 5N4W, 5T35). Residues marked in red were identified in hybrid capture analysis. See also Fig. 1 and Supplementary Dataset. (e) Number of spontaneous degrader resistance alterations in the substrate receptor (CRBN, VHL, colored) binned by their distance to the degrader-binding site. See also Fig. 1d and Supplementary Dataset.
Extended Data Fig. 2 Characterization of DMS Libraries and AT7.
(a) Pie charts depicting the distribution of different alterations identified by sequencing the mutational scanning libraries for CRBN (top) and VHL (bottom). (b) Stacked bar graphs and density distributions of residue wise normalized abundance of mutants identified in the DMS libraries for VHL (top) and CRBN (bottom). (c) Chemical structure comparison of the degraders AT1 and AT7. (d) Dose-resolved, normalized viability after 3 d treatment with MZ-1, macroPROTAC-1, cis MZ-1 (a non VHL binding control of MZ-1 or AT7 in MV4;11 cells. Mean ± s.e.m.; n = 3 independent treatments. (e) Dose-resolved, normalized viability after 3 d treatment (AT7) in RKO VHL−/− cells with over-expression of VHLWT. Mean ± s.e.m.; n = 3 independent treatments. (f) Protein levels in HeLa cells treated with MZ-1 or AT7 (18 h, indicated concentration). Representative images of n = 2 independent measurements. (g) Protein levels in RKO VHL−/− cells with over-expression of VHLWT treated with DMSO or AT7 (60 nM, 2 h). Representative images of n = 2 independent measurements. (h) Dose-resolved, normalized viability after 4 d treatment (ACBI1) and 3 d treatment (ARV-771, MZ-1, macroPROTAC-1) in RKO VHL−/− cells with over-expression of VHLWT. Mean ± s.e.m.; n = 3 independent treatments.
Extended Data Fig. 3 DMS Robustly Locates Functional Hotspots of General Relevance in the Degrader Binding Pocket.
(a) Scatter plot depicting log2 fold-enrichment between different batch mutational scanning resistance measurements of VHL (500 nM ARV-771) or CRBN mutations (500 nM dBET6) normalized to DMSO after 7-day treatment. The rank-based measure of association was estimated via Spearman’s rho statistic and reported P-values were calculated via asymptotic two-sided t approximation without adjustments for multiple comparisons. (b) Stacked bar graphs of log2 fold-enrichment of VHL mutants normalized to DMSO treated with the indicated concentrations of ARV-771 for 7 days. n = 2 independent measurements. (c) Dose-resolved, normalized viability after 3 d treatment with dBET6, CC-90009, dBET57 or CC-885 in RKO CRBN−/− cells with over-expression of CRBNWT. Mean ± s.e.m.; n = 3 independent treatments. (d) Surface structure of CRBN bound by dBET6 (PDB 6BOY). Median log2 fold-enrichment of all CRBN mutations over DMSO across 4 degrader treatments (see Fig. 2d) is mapped in purple to dark grey onto positions mutated in the CRBN library.
Extended Data Fig. 4 Functional VHL Hotspots Show Neo-Substrate Specific Resistance and Sensitivity to PROTACs.
(a and b) Dose-resolved, normalized viability after 4 d treatment (ACBI1) and 3 d treatment (MZ-1) in RKO VHL−/− cells with over-expression of VHLWT, VHLR69G or VHLN67Q. Mean ± s.e.m.; n = 3 independent treatments. (c and d) Depiction of clonogenic assays via crystal violet staining. RKO VHL−/− cells with over-expression of VHLWT, VHLR69G, VHLN67R or VHLN67Q were treated for 10 days at EC90 of the degrader (2.5 uM ACBI1, 50 nM ARV-771, 75 nM MZ-1). (e) Protein levels in RKO VHL−/− cells with over-expression of VHLWT or VHLN67Q treated with DMSO, MZ-1 (75 nM, 2 h), ARV-771 (50 nM, 2 h) or ACBI1 (2.5 uM, 4 h). Representative images of n = 2 independent measurements. (f) Cocrystal structure of MZ-1 in a ternary complex with VHL-ElonginC-ElonginB and BRD4BD2 (PDB: 5T35). (g) Heatmap depicting differential log2 fold-enrichment of the VHLH110 mutations normalized to DMSO after treatment with ARV-771 (500 nM, 7d). n = 2 independent measurements. (h) Dose-resolved, normalized viability after 3d treatment AT7 (top), macroPROTAC-1 (bottom, left) or ARV-771 (bottom, right) in RKO VHL−/− cells with over-expression of VHLWT or VHLH110L. Mean ± s.e.m.; n = 3 independent treatments. (i) Protein levels in RKO VHL−/− cells with over-expression of VHLWT or VHLH110L treated with DMSO or AT7 (60 nM, 2 h). Representative images of n = 2 independent measurements. (j) Overlay of cocrystal structures of AT2 (grey, purple, blue) and MZ1 (black, PDB:5T35) in a ternary complex with VHL-ElonginC-ElonginB and BRD4BD2 showing a lateral shift of BRD4BD2. (k) Cocrystal structure of AT2 in a ternary complex with VHL-ElonginC-ElonginB and BRD4BD2. See Fig. 3. (l) Dose-resolved, normalized viability after 4 d treatment (ACBI1) and 3 d treatment (MZ-1, ARV-771) in RKO VHL-/- cells with over-expression of VHLWT or VHLY112C. Mean ± s.e.m.; n = 3 independent treatments.
Extended Data Fig. 5 VHLP71 is a Functional Hotspot for Degrader Specific Resistance.
(a) Heatmap depicting differential log2 fold-enrichment of the VHLP71 mutations normalized to DMSO between treatment with ARV-771 (500 nM, 7d) and macroPROTAC-1 (2 uM, 7d). n = 2 independent measurements. (b) Depiction (left) and quantification (right) of clonogenic assays via crystal violet staining. RKO VHL−/− cells with over-expression of VHLWT or VHLP71I were treated for 10 days at EC90 of the degrader (50 nM ARV-771, 75 nM MZ-1, 1 uM macroPROTAC-1).
Extended Data Fig. 6 Functional CRBN Hotspots Show Degrader Selectivity.
(a, c and f) Depiction of clonogenic assays via crystal violet staining. RKO CRBN−/− cells with over-expression of CRBNWT, CRBNE377K, CRBNN351D or CRBNH397D were treated for 10 days with DMSO, 30 nM dBET6, 60 nM CC-90009, 480 nM dBET57 or the indicated concentration of THAL-SNS-032. (b and g) Dose-resolved, normalized viability after 3 d treatment with THAL-SNS-032, dBET6 or CC-90009 in RKO CRBN−/− cells with over-expression of CRBNWT, CRBNN351D, CRBNH397Y or CRBNH57D. Mean ± s.e.m.; n = 3 independent treatments. (d) Protein levels in RKO CRBN−/− cells with over-expression of CRBNWT or CRBNE377K treated with DMSO, CC-90009 (50 nM, 6 h) or dBET6 (15 nM, 2 h). Representative images of n = 2 independent measurements. (e) Heatmap depicting differential log2 fold-enrichment of CRBNH397 mutations normalized to DMSO with dBET57 treatment (500 nM, 7d). n = 3 independent measurements. (h) Quantification of clonogenic assays via crystal violet extraction and measurement of absorption at 590 nM. RKO CRBN−/− cells with over-expression of CRBNWT or CRBNH57D were treated for 10 days with DMSO, 30 nM dBET6, 60 nM CC-90009, 480 nM dBET57 or 0.6 nM CC-885. See also Fig. 5.
Supplementary information
Supplementary Information
Supplementary Tables 1–4, Supplementary Figures 1–3, Supplementary Note
Supplementary Data
Hybrid-capture results.
Source data
Source Data Fig. 3
Unprocessed western blot images related to Fig. 3.
Source Data Fig. 4
Unprocessed western blot images related to Fig. 4.
Source Data Fig. 5
Unprocessed western blot images related to Fig. 5.
Source Data Extended Data Fig. 2
Unprocessed western blot images related to Extended Data Fig. 2.
Source Data Extended Data Fig. 4
Unprocessed western blot images related to Extended Data Fig. 4.
Source Data Extended Data Fig. 6
Unprocessed western blot images related to Extended Data Fig. 6.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hanzl, A., Casement, R., Imrichova, H. et al. Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders. Nat Chem Biol 19, 323–333 (2023). https://doi.org/10.1038/s41589-022-01177-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01177-2
This article is cited by
-
DCAF1-based PROTACs with activity against clinically validated targets overcoming intrinsic- and acquired-degrader resistance
Nature Communications (2024)
-
Discovery of a Drug-like, Natural Product-Inspired DCAF11 Ligand Chemotype
Nature Communications (2023)
-
Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2
Nature Communications (2023)
-
Identifying degrader hotspots
Nature Reviews Drug Discovery (2023)
-
The rise of targeting chimeras (TACs): next-generation medicines that preempt cellular events
Medicinal Chemistry Research (2023)