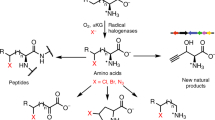
Abstract
FeII/α-ketoglutarate (FeII/αKG)-dependent enzymes offer a promising biocatalytic platform for halogenation chemistry owing to their ability to functionalize unactivated C–H bonds. However, relatively few radical halogenases have been identified to date, limiting their synthetic utility. Here, we report a strategy to expand the palette of enzymatic halogenation by engineering a reaction pathway rather than substrate selectivity. This approach could allow us to tap the broader class of FeII/αKG-dependent hydroxylases as catalysts by their conversion to halogenases. Toward this goal, we discovered active halogenases from a DNA shuffle library generated from a halogenase–hydroxylase pair using a high-throughput in vivo fluorescent screen coupled to an alkyne-producing biosynthetic pathway. Insights from sequencing halogenation-active variants along with the crystal structure of the hydroxylase enabled engineering of a hydroxylase to perform halogenation with comparable activity and higher selectivity than the wild-type halogenase, showcasing the potential of harnessing hydroxylases for biocatalytic halogenation.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Accession codes for proteins in this study are as follows: Hal, WP_122981682; Hydrox, WP_107105619. The PDB accession code for the structure of the hydroxylase from Streptomyces sp. MBT76 (Hydrox, WP_107105619) is 7JSD. The PDB accession code for BesD is 6NIE. NMR data and shuffle sequencing data are also provided. Any additional datasets from the supplemental information generated during and/or analyzed during the current study are available from the corresponding author on request. Source data are provided with this paper.
Code availability
The code used to analyze the shuffle data is included in the Supplementary Information.
References
Koeller, K. M. & Wong, C.-H. Enzymes for chemical synthesis. Nature 409, 232–240 (2001).
Sharma, S. V. et al. Living GenoChemetics by hyphenating synthetic biology and synthetic chemistry in vivo. Nat. Commun. 8, 229 (2017).
Prier, C. K., Zhang, R. K., Buller, A. R., Brinkmann-Chen, S. & Arnold, F. H. Enantioselective, intermolecular benzylic C–H amination catalysed by an engineered iron-haem enzyme. Nat. Chem. 9, 629–634 (2017).
Latham, J., Brandenburger, E., Shepherd, S. A., Menon, B. R. K. & Micklefield, J. Development of halogenase enzymes for use in synthesis. Chem. Rev. 118, 232–269 (2018).
Gkotsi, D. S., Dhaliwal, J., McLachlan, M. M., Mulholand, K. R. & Goss, R. J. Halogenases: powerful tools for biocatalysis (mechanisms, applications, and scope). Curr. Opin. Chem. Biol. 43, 119–126 (2018).
Gkotsi, D. S. et al. A marine viral halogenase that iodinates diverse substrates. Nat. Chem. 11, 1091–1097 (2019).
Payne, J. T. et al. Enantioselective desymmetrization of methylenedianilines via enzyme-catalyzed remote halogenation. J. Am. Chem. Soc. 140, 546–549 (2018).
Agarwal, V. et al. Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem. Rev. 117, 5619–5674 (2017).
Vaillancourt, F. H., Yeh, E., Vosburg, D. A., O’Connor, S. E. & Walsh, C. T. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature 436, 1191–1194 (2005).
Galonić, D. P., Vaillancourt, F. H. & Walsh, C. T. Halogenation of unactivated carbon centers in natural product biosynthesis: trichlorination of leucine during barbamide biosynthesis. J. Am. Chem. Soc. 128, 3900–3901 (2006).
Ueki, M. et al. Enzymatic generation of the antimetabolite γ,γ-dichloroaminobutyrate by NRPS and mononuclear iron halogenase action in a streptomycete. Chem. Biol. 13, 1183–1191 (2006).
Neumann, C. S. & Walsh, C. T. Biosynthesis of (−)-(1 S, 2 R)-allocoronamic acyl thioester by an FeII-dependent halogenase and a cyclopropane-forming flavoprotein. J. Am. Chem. Soc. 130, 14022–14023 (2008).
Jiang, W. et al. Biosynthetic chlorination of the piperazate residue in kutzneride biosynthesis by KthP. Biochemistry 50, 6063–6072 (2011).
Khare, D. et al. Conformational switch triggered by α-ketoglutarate in a halogenase of curacin A biosynthesis. Proc. Natl Acad. Sci. USA 107, 14099–14104 (2010).
Flatt, P. M. et al. Characterization of the initial enzymatic steps of barbamide biosynthesis. J. Nat. Prod. 69, 938–944 (2006).
Pratter, S. M. et al. More than just a halogenase: modification of fatty acyl moieties by a trifunctional metal enzyme. ChemBioChem 15, 567–574 (2014).
Hillwig, M. L. & Liu, X. A new family of iron-dependent halogenases acts on freestanding substrates. Nat. Chem. Biol. 10, 6–10 (2014).
Hillwig, M. L., Zhu, Q., Ittiamornkul, K. & Liu, X. Discovery of a promiscuous non-heme iron halogenase in ambiguine alkaloid biogenesis: implication for an evolvable enzyme family for late-stage halogenation of aliphatic carbons in small molecules. Angew. Chem. Int. Ed. Engl. 55, 5780–5784 (2016).
Kim, C. Y. et al. The chloroalkaloid (−)-acutumine is biosynthesized via a Fe(II)- and 2-oxoglutarate-dependent halogenase in Menispermaceae plants. Nat. Commun. 11, 1867 (2020).
Zhao, C., et al. An Fe2+- and α-ketoglutarate-dependent halogenase acts on nucleotide substrates. Angew. Chem. Int. Ed. Engl. 59, 9478–9484 (2020).
Marchand, J. A. et al. Discovery of a pathway for terminal-alkyne amino acid biosynthesis. Nature 567, 420–424 (2019).
Islam, M. S., Leissing, T. M., Chowdhury, R., Hopkinson, R. J. & Schofield, C. J. 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem. 87, 585–620 (2018).
Schofield, C & Hausinger, R. 2-Oxoglutarate-Dependent Oxygenases (The Royal Society of Chemistry, London, 2015).
Blasiak, L. C., Vaillancourt, F. H., Walsh, C. T. & Drennan, C. L. Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature 440, 368–371 (2006).
Galonić, D. P., Barr, E. W., Walsh, C. T., Bollinger, J. M. & Krebs, C. Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nat. Chem. Biol. 3, 113–116 (2007).
Matthews, M. L. et al. Substrate positioning controls the partition between halogenation and hydroxylation in the aliphatic halogenase, SyrB2. Proc. Natl Acad. Sci. USA 106, 17723–17728 (2009).
Wong, S. D. et al. Elucidation of the Fe(IV) = O intermediate in the catalytic cycle of the halogenase SyrB2. Nature 499, 320–323 (2013).
Mitchell, A. J. et al. Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5. Nat. Chem. Biol. 12, 636–640 (2016).
Martinie, R. J. et al. Experimental correlation of substrate position with reaction outcome in the aliphatic halogenase, SyrB2. J. Am. Chem. Soc. 137, 6912–6919 (2015).
Chaplin, V. D. et al. Chloride supports O2 activation in the D201G facial triad variant of factor-inhibiting hypoxia inducible factor, an α-ketoglutarate dependent oxygenase. Inorg. Chem. 57, 12588–12595 (2018).
Bollinger, J. M., et al. in 2-Oxoglutarate-Dependent Oxygenases Ch. 3 (The Royal Society of Chemistry, London, 2015).
Srnec, M. & Solomon, E. I. Frontier molecular orbital contributions to chlorination versus hydroxylation selectivity in the non-heme iron halogenase SyrB2. J. Am. Chem. Soc. 139, 2396–2407 (2017).
Gorres, K. L., Pua, K. H. & Raines, R. T. Stringency of the 2-His–1-Asp active-site motif in prolyl 4-hydroxylase. PLoS ONE 4, e7635 (2009).
Grzyska, P. K., Müller, T. A., Campbell, M. G. & Hausinger, R. P. Metal ligand substitution and evidence for quinone formation in taurine/α-ketoglutarate dioxygenase. J. Inorg. Biochem. 101, 797–808 (2007).
Hewitson, K. S. et al. Evidence that two enzyme-derived histidine ligands are sufficient for iron binding and catalysis by factor inhibiting HIF (FIH). J. Biol. Chem. 283, 25971–25978 (2008).
Mitchell, A. J. et al. Structure-guided reprogramming of a hydroxylase to halogenate its small molecule substrate. Biochemistry 56, 441–444 (2017).
Papadopoulou, A., et al. Re-programming and optimization of a l-proline cis-4-hydroxylase for the cis-3-halogenation of its native substrate. ChemCatChem 13, 3914–3919 (2021).
Neugebauer, M. E. et al. A family of radical halogenases for the engineering of amino-acid-based products. Nat. Chem. Biol. 15, 1009–1016 (2019).
Luo, L. et al. An assay for Fe(II)/2-oxoglutarate-dependent dioxygenases by enzyme-coupled detection of succinate formation. Anal. Biochem. 353, 69–74 (2006).
Guo, A. C. et al. ECMDB: the E. coli metabolome database. Nucleic Acids Res. 41, D625–D630 (2013).
Iyer, S. R., Chaplin, V. D., Knapp, M. J. & Solomon, E. I. O2 activation by nonheme FeII α-ketoglutarate-dependent enzyme variants: elucidating the role of the facial triad carboxylate in FIH. J. Am. Chem. Soc. 140, 11777–11783 (2018).
Hangasky, J. A., Taabazuing, C. Y., Valliere, M. A. & Knapp, M. J. Imposing function down a (cupin)-barrel: secondary structure and metal stereochemistry in the αKG-dependent oxygenases. Metallomics 5, 287 (2013).
Mehmood, R., Vennelakanti, V. & Kulik, H. J. Spectriscopically guided simulations reveal distinct strategies for positioning substrates to achieve selectivity in nonheme Fe(II)/α-ketoglutarate-dependent halogenases. ACS Catal. 11, 12394–12408 (2021).
Zhu, X., Shieh, P., Su, M., Bertozzi, C. R. & Zhang, W. A fluorogenic screening platform enables directed evolution of an alkyne biosynthetic tool. Chem. Commun. 52, 11239–11242 (2016).
Varazo, K., Droumaguet, C. L., Fullard, K. & Wang, Q. Metal ion detection using a fluorogenic ‘click’ reaction. Tetrahedron Lett. 50, 7032–7034 (2009).
Shieh, P. et al. CalFluors: a universal motif for fluorogenic azide probes across the visible spectrum. J. Am. Chem. Soc. 137, 7145–7151 (2015).
Saban, E. et al. The second coordination sphere of FIH controls hydroxylation. Biochemistry 50, 4733–4740 (2011).
Huang, Y., Niu, B., Gao, Y., Fu, L. & Li, W. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 (2010).
Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004).
Larsson, A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278 (2014).
Jones, D. T., Taylor, W. R. & Thornton, J. M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 (1992).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Bryksin, A. & Matsumura, I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48, 463–465 (2010).
Gasteiger, E. et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Pettersen, E. F. et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
This work was supported by DOE/LBNL DEAC02-05CH11231, FWP CH030201. M.E.N. acknowledges the support of a National Science Foundation Graduate Research Fellowship. E.N.K. acknowledges the support of a National Institutes of Health NRSA Training Grant (1 T32 GMO66698). J.A.M. acknowledges the support of a UC Berkeley Chancellor’s Fellowship, Howard Hughes Medical Institute Gilliam Fellowship and National Institutes of Health NRSA Training Grant (1 T32 GMO66698). X-ray data were collected at the Advanced Light Source Beamline 8.3.1, which is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives (MR-15- 328599), the National Institutes of Health (R01 GM124149 and P30 GM124169), Plexxikon and the Integrated Diffraction Analysis Technologies program of the US Department of Energy Office of Biological and Environmental Research. The Advanced Light Source is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy under contract number DEAC02-05CH11231, Office of Basic Energy Sciences. The funds for the 900 MHz NMR spectrometer housed in the QB3 Institute in Stanley Hall at University of California, Berkeley, were kindly provided by the NIH (GM68933).
Author information
Authors and Affiliations
Contributions
M.E.N. performed enzyme characterization experiments, library generation, screen development, library analysis and assisted with protein crystallography. E.N.K. performed protein crystallography and enzyme characterization experiments. J.A.M. and D.C.M. contributed to screen development. J.G.P. performed NMR experiments. N.A.S. assisted with enzyme characterization experiments. M.C.Y.C. designed experiments and administered the project. M.E.N. and M.C.Y.C. wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemical Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Divergent mechanisms of FeII/αKG-dependent halogenases and hydroxylases.
FeII/αKG-dependent halogenases and hydroxylases are mechanistically related. Upon oxygen binding, both enzymes decarboxylate αKG to generate a FeIV-oxo intermediate for abstracting a hydrogen atom from the substrate. However, differences in the primary coordination sphere of iron contribute to the subsequent reaction outcome. In hydroxylases, iron is coordinated by Asp or Glu, and hydroxyl rebound with the substrate radical leads to hydroxylation. In contrast, halogenases lack Asp or Glu in the primary coordination sphere and instead have Gly or Ala, leaving space for chloride to coordinate to Fe. Although rebound of either the halogen or the hydroxyl is possible, halogenases have evolved to favor halogen rebound.
Extended Data Fig. 2 Sequence alignment of BesD, Hal, and Hydrox.
(A) Sequence identity matrix for the halogenases from S. cattleya (BesD) and A. teichomyceticus (Hal, WP_122981682), and the hydroxylase from S. sp. MBT76 (Hydrox, WP_107105619). (B) Sequence alignment of BesD, Hal, and Hydrox highlighting the HXG and HXD motifs.
Extended Data Fig. 3 Steady-state kinetic analysis of Hal and Hydrox.
The amino acid lysine was used as a substrate. Data are mean ± sd (n = 3 technical replicates). Table contains kcat, KM, and kcat/KM calculated by non-linear curve fitting to the Michaelis-Menten equation. Data in the table (kinetic parameters) are mean ± s.e. Error in kcat/KM is obtained by propagation from the individual kinetic terms.
Extended Data Fig. 4 Results of screening the shuffle library with CuAAC.
Cells expressing BesB, BesC, and a member of the shuffle library are grown in M9 media supplemented with 0.5 mM lysine for two days. Controls are present on each plate as follows: Hydrox (A1-4), Hydrox D144G (A5-8), Hal (A9-12). After two days of expression, cells were pelleted by centrifugation and the supernatant was transferred to a fresh plate and mixed 1:1 with CuAAC solution to yield the final reaction mixture (CalFluor 488 azide (1 μM), copper (II) sulfate (0.5 mM), BTTAA (0.1 mM), and sodium ascorbate (5 mM)). Following incubation in the dark for 15 min, the fluorescence was measured using a SynergyMx Microplate Reader (BioTek) at room temperature (λex = 485 nm, λem = 528 nm). Values for each well are reported relative to the average of wells A1-4 (Hydrox) on each plate.
Extended Data Fig. 5 Purification and LC/MS analysis of Hal G144D I151N N225V.
(A) SDS-PAGE gel of purified Hal G144D I151N N225V. Protein purification was performed once. (B) Extracted ion chromatograms of LC/MC analysis of chlorolysine (m/z = 181.0738) and hydroxylysine (m/z = 163.1077) produced by Hal, Hydrox, and Hal G144D I151N N225V. Reactions contained Fe, lysine, NaCl, αKG, and purified enzyme variants and proceeded for 25 min before quenching in methanol containing 1% (v/v) formic acid to precipitate protein prior to LC/MS analysis. The Hal triple mutant produces no chlorolysine as expected given the G144D mutation likely abolishes chloride binding. A 3-fold increase in hydroxylysine production is observed in Hal G144D I151N N225V the compared to wild-type Hal, which remains 1.5-fold lower than wild-type Hydrox.
Extended Data Fig. 6 Purification and LC/MS analysis of HalI151 variants.
(A) SDS-PAGE gel of purified Hal and Hydrox variants. Lane 8, Hal I151V; 9, Hal I151A; 10, Hal I151N. Protein purification was performed once. (B) LC/MS analysis of chlorolysine (red, m/z = 181.0738) and hydroxylysine (black, m/z = 163.1077) produced by Hal, Hal I151V, Hal I151A, Hal I151N, and Hydrox. Reactions containing Fe, lysine, NaCl, αKG, and purified enzyme variants proceeded for 25 min before quenching in methanol containing 1% (v/v) formic acid to precipitate protein prior to LC/MS analysis. Mean and standard error are shown for n = 3 technical replicates. Selectivity values are reported in Supplementary Table 3.
Extended Data Fig. 7 Designing a chimera based on the shuffle library.
To design an enzyme which captures the key sequence motifs required for halogenation, a chimeric protein was engineered based on the shuffle results. (A) Conservation of halogenase residues that were conserved in library members capable of performing halogenation. Residues that were conserved in 100% of the halogenating hits were grafted into the sequence of the hydroxylase. All the residues that differ between the Chi-14 and Hydrox are listed below the sequence map. Key primary and secondary sphere residues are highlighted in green. (B) Sequence alignment of Hydrox (blue), Hal (red), and the engineered Chi-14.
Extended Data Fig. 8 Steady-state kinetic analysis of Hydrox triple mutant and Chi-14.
The amino acid lysine was used as a substrate. Data are mean ± sd (n = 3 technical replicates). Table contains kcat, KM, and kcat/KM calculated by non-linear curve fitting to the Michaelis-Menten equation. Data in the table (kinetic parameters) are mean ± s.e. Error in kcat/KM is obtained by propagation from the individual kinetic terms.
Extended Data Fig. 9 Total turnover number (TTN) and succinate coupling ratios for Hal, Hydrox, Hydrox D144G N151I V225N, and Chi-14.
Reactions (50 µL) contained l-lysine (4-7 mM), sodium αKG (10 mM), sodium ascorbate (5 mM), (NH4)2Fe(SO4)2 · 6H2O (1 mM), and sodium chloride (10 mM) in 100 mM MOPS buffer (pH 7.5). Reactions were initiated by addition of purified Hal or Hydrox variants (12.5 or 25 µM final concentration) and allowed to proceed for 24 hours at room temperature before quenching in 2.5 vol of methanol with 1% (v/v) formic acid. Following centrifugation at 20,000 × g for 20 min (at 4 °C) to remove precipitated protein, lysine and succinate concentrations were analyzed by LC/MS. (A) Standard curve for lysine quantification by LC/MS. Mean and standard error are shown for n = 3 technical replicates. (B) Lysine turnover for Hydrox, Hal, Hydrox D144G N151I V225N, and Chi-14. Mean and standard error are shown for n = 3 technical replicates. (C) Standard curve for succinate quantification by LC/MS. Mean and standard error are shown for n = 3 technical replicates. (D) Succinate turnover for Hydrox, Hal, Hydrox D144G N151I V225N, and Chi-14. Mean and standard error are shown for n = 3 technical replicates.
Extended Data Fig. 10 Purification and LC/MS analysis of Hydrox and Chi-14 variants.
(A) SDS-PAGE gel of purified Hydrox D144G N151I V225N (triple mutant, HydroxTM) and Chi-14 variants. Lane 11, Chi-14 L150F; 12, Chi-14 ST218-219GAV; 13, Chi-14 I222M; 14, Chi-14 M225V T226S; 15, Chi-14 A228S; 16, Chi-14 K230E E234G D236V; 17, HydroxTM F150L; 18, HydroxTM GAV218/219 to ST; 19, HydroxTM M223I; 20, HydroxTM V226M S227T; 21, HydroxTM S229A; 22, HydroxTM E231K G235E V237D. Proteins were purified once. (B) LC/MS analysis of chlorolysine (red, m/z = 181.0738) and hydroxylysine (black, m/z = 163.1077) produced by Hydrox and Chi-14 variants. The variants 11-22 correspond to the purified proteins in (A). An asterisk and the corresponding dotted line provide points of comparison for Chi-14 mutants that diminish activity relative to Chi-14 or Hydrox D144G N151I V225N (HydroxTM) mutants that increase activity relative to the triple mutant, Hydrox D144G N151I V225N. Reactions containing Fe, lysine, NaCl, αKG, and purified enzyme variants proceeded for 25 min before quenching in methanol containing 1% (v/v) formic acid to precipitate protein prior to LC/MS analysis. Mean and standard error are shown for n = 3 technical replicates. Selectivity values are reported in Supplementary Table 3.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Tables 1–6, Notes 1–3.
Supplementary Data 1
NMR analysis of 4-hydroxylysine produced by Hydrox.
Supplementary Data 2
NMR analysis of 2-amino-4-hydroxy-ε-lactam produced by PfHalA.
Supplementary Data 3
NMR analysis of 2-amino-4-hydroxy-ε-lactam produced by Chi-14.
Supplementary Data 4
Raw sequencing data used to generate Fig. 4.
Source data
Source Data Fig. 1
LC–MS data.
Source Data Fig. 3
Fluorescence plate reader data.
Source Data Fig. 4
Residue conservation scatter data.
Source Data Fig. 5
LC–MS data, integrated EIC.
Source Data Extended Data Fig. 3
Michaelis–Menten kinetics data.
Source Data Extended Data Fig. 4
Fluorescence plate reader data, CuAAC.
Source Data Extended Data Fig. 5
LC–MS data.
Source Data Extended Data Fig. 6
LC–MS data, integrated EIC.
Source Data Extended Data Fig. 8
Michaelis–Menten kinetics data.
Source Data Extended Data Fig. 9
Succinate coupling data.
Source Data Extended Data Fig. 10
LC–MS data, integrated EIC.
Rights and permissions
About this article
Cite this article
Neugebauer, M.E., Kissman, E.N., Marchand, J.A. et al. Reaction pathway engineering converts a radical hydroxylase into a halogenase. Nat Chem Biol 18, 171–179 (2022). https://doi.org/10.1038/s41589-021-00944-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00944-x
This article is cited by
-
Engineering non-haem iron enzymes for enantioselective C(sp3)–F bond formation via radical fluorine transfer
Nature Synthesis (2024)
-
Enabling technology and core theory of synthetic biology
Science China Life Sciences (2023)