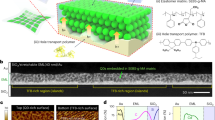
Abstract
The field of engineered living materials aims to construct functional materials with desirable properties of natural living systems. A recent study demonstrated the programmed self-assembly of bacterial populations by engineered adhesion. Here we use this strategy to engineer self-healing living materials with versatile functions. Bacteria displaying outer membrane-anchored nanobody–antigen pairs are cultured separately and, when mixed, adhere to each other to enable processing into functional materials, which we term living assembled material by bacterial adhesion (LAMBA). LAMBA is programmable and can be functionalized with extracellular moieties up to 545 amino acids. Notably, the adhesion between nanobody–antigen pairs in LAMBA leads to fast recovery under stretching or bending. By exploiting this feature, we fabricated wearable LAMBA sensors that can detect bioelectrical or biomechanical signals. Our work establishes a scalable approach to produce genetically editable and self-healable living functional materials that can be applied in biomanufacturing, bioremediation and soft bioelectronics assembly.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that all the source data processed for figures generation in this study are available within the paper, source data file and the supplementary data files. Any additional information is available upon reasonable request. Source data are provided with this paper.
Code availability
The code that supports the findings of this study are available from the corresponding author upon reasonable request.
References
Chen, A. Y., Zhong, C. & Lu, T. K. Engineering living functional materials. ACS Synth. Biol. 4, 8–11 (2015).
Nguyen, P. Q., Courchesne, N. M. D., Duraj-Thatte, A., Praveschotinunt, P. & Joshi, N. S. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 30, e1704847 (2018).
Gilbert, C. & Ellis, T. Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 8, 1–15 (2019).
Tang, T. C. et al. Materials design by synthetic biology. Nat. Rev. Mater. 6, 332–350 (2021).
Chen, A. Y. et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 13, 515–523 (2014).
Praveschotinunt, P. et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 10, 5580 (2019).
Moser, F., Tham, E., González, L. M., Lu, T. K. & Voigt, C. A. Light-controlled, high-resolution patterning of living engineered bacteria onto textiles, ceramics, and plastic. Adv. Funct. Mater. 29, 11 (2019).
Nguyen, P. Q., Botyanszki, Z., Tay, P. K. & Joshi, N. S. Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 5, 4945 (2014).
Huang, J. et al. Programmable and printable Bacillus subtilis biofilms as engineered living materials. Nat. Chem. Biol. 15, 34–41 (2019).
Zhong, C. et al. Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nat. Nanotechnol. 9, 858–866 (2014).
Evans, M. L. & Chapman, M. R. Curli biogenesis: order out of disorder. Biochim. Biophys. Acta 1843, 1551–1558 (2014).
Barnhart, M. M. & Chapman, M. R. Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 (2006).
Petrova, O. E. & Sauer, K. Sticky situations: key components that control bacterial surface attachment. J. Bacteriol. 194, 2413–2425 (2012).
Ahn, B. K., Lee, D. W., Israelachvili, J. N. & Waite, J. H. Surface-initiated self-healing of polymers in aqueous media. Nat. Mater. 13, 867–872 (2014).
Huang, Y. et al. A self-healable and highly stretchable supercapacitor based on a dual crosslinked polyelectrolyte. Nat. Commun. 6, 10310 (2015).
Glass, D. S. & Riedel-Kruse, I. H. A synthetic bacterial cell–cell adhesion toolbox for programming multicellular morphologies and patterns. Cell 174, 649–658.e616 (2018).
Tsoi, R., Dai, Z. & You, L. Emerging strategies for engineering microbial communities. Biotechnol. Adv. 37, 107372 (2019).
Hays, S. G., Patrick, W. G., Ziesack, M., Oxman, N. & Silver, P. A. Better together: engineering and application of microbial symbioses. Curr. Opin. Biotechnol. 36, 40–49 (2015).
Han, M. J. & Lee, S. H. An efficient bacterial surface display system based on a novel outer membrane anchoring element from the Escherichia coli protein YiaT. FEMS Microbiol. Lett. 362, 1–7 (2015).
Rollié, S., Mangold, M. & Sundmacher, K. Designing biological systems: systems engineering meets synthetic biology. Chem. Eng. Sci. 69, 1–29 (2012).
Behrendorff, J. B. & Gillam, E. M. Prospects for applying synthetic biology to toxicology: future opportunities and current limitations for the repurposing of cytochrome P450 systems. Chem. Res. Toxicol. 30, 453–468 (2017).
Tsoi, R. et al. Metabolic division of labor in microbial systems. Proc. Natl Acad. Sci. USA 115, 2526–2531 (2018).
Thommes, M., Wang, T., Zhao, Q., Paschalidis, I. C. & Segrè, D. Designing metabolic division of labor in microbial communities. mSystems 4, e00263–18 (2019).
Kim, D. H. et al. Epidermal electronics. Science 333, 838–843 (2011).
Schiavone, G. & Lacour, S. P. Conformable bioelectronic interfaces: mapping the road ahead. Sci. Transl. Med. 11, https://doi.org/10.1126/scitranslmed.aaw5858 (2019).
Liu, Y. et al. Soft and elastic hydrogel-based microelectronics for localized low-voltage neuromodulation. Nat. Biomed. Eng. 3, 58–68 (2019).
Chortos, A., Liu, J. & Bao, Z. Pursuing prosthetic electronic skin. Nat. Mater. 15, 937–950 (2016).
Jiheong Kang, J. B.-H. T. Z. B. Self-healing soft electronics. Nat. Electron. 2, 144–150 (2019).
Cai, G. et al. Extremely stretchable strain sensors based on conductive self-healing dynamic cross-links hydrogels for human-motion detection. Adv. Sci. 4, 1600190 (2017).
Wu, J. et al. Highly stretchable and transparent thermistor based on self-healing double network hydrogel. ACS Appl. Mater. Interfaces 10, 19097–19105 (2018).
Zhu, M., et al. Towards optimizing electrode configurations for silent speech recognition based on high-density surface electromyography. J. Neural Eng. 18, 016005 (2020).
Jiang, N., Wang, L., Huang, Z. & Li, G. Mapping responses of lumbar paravertebral muscles to single-pulse cortical TMS using high-density surface electromyography. IEEE Trans. Neural Syst. Rehabil. Eng. 29, 831–840 (2021).
Yu, Y. et al. Simple spinning of heterogeneous hollow microfibers on chip. Adv. Mater. 28, 6649–6655 (2016).
Acknowledgements
We thank D.S. Glass for sharing plasmids; X. Shen for sharing strains and instructive comments; L. Jiang for insightful comments on trehalose synthesis experiments; F. Jin and S. Huang for assistance in microscopy and microfluidic instruments setup; C. Liu and W. Liu for help in strain engineering; the Testing Technology Center of Materials and Devices and the Tsinghua Shenzhen International Graduate School for TEM instrument usage. This study was partially supported by National Key Research and Development Program of China (grant nos. 2018YFA0903000 and 2020YFA0908100 to Z.D.: these two grants provide equal support), National Natural Science Foundation of China National Natural Science Foundation of China grant nos. 81927804 (Z.L.) and 32071427 (Z.D.). Shenzhen Science and Technology Program grant no. KQTD20180413181837372 (Z.D.).
Author information
Authors and Affiliations
Contributions
B.C. designed and performed the experiments, interpreted the results and wrote the paper. W.K. assisted in experimental setup and data interpretation. J.S. designed and performed the experiments, interpreted the results and revised the paper. R.Z. performed the experiments, interpreted the results and revised the paper. Yue Y. and A.X. assisted in experimental setup and data interpretation of microfluidic and microscopy. M.Y., M.W. and J.H. assisted in performing experiments and experimental setup in electronic circuit assembly, 3D printing and rheology measurement. Y.C., L.T. and Q.T. assisted in performing experiments and experimental setup in living fiber generation and EMG measurement. Yin Y., G.L. and L.Y. assisted in research design, experimental setup, data interpretation and paper revisions. Z.L. conceived the research, assisted in the experimental design, results interpretation and paper revisions. Z.D. conceived the research, designed the experiments, interpreted the results and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemical Biology thanks Neel Joshi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1-21, Tables 1–4 and Videos 1–3.
Supplementary Video 1
Growth of the LAMBA fiber observed by microscopy.
Supplementary Video 2
LAMBA sensor monitored the cyclic finger joint motion.
Supplementary Video 3
The traditional sensor made of gold film failed to monitor the finger joint bending due to the limitation of the stretchability.
Source data
Source Data Fig. 1
One file for Fig. 1, containing all source data.
Source Data Fig. 2
One file for Fig. 2, containing all source data.
Source Data Fig. 3
One file for Fig. 3, containing all source data.
Source Data Fig. 4
One file for Fig. 4, containing all source data.
Rights and permissions
About this article
Cite this article
Chen, B., Kang, W., Sun, J. et al. Programmable living assembly of materials by bacterial adhesion. Nat Chem Biol 18, 289–294 (2022). https://doi.org/10.1038/s41589-021-00934-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00934-z
This article is cited by
-
Synthetic microbiology in sustainability applications
Nature Reviews Microbiology (2024)
-
De novo engineering of a bacterial lifestyle program
Nature Chemical Biology (2023)
-
A change of phase
Nature Chemical Biology (2023)
-
Synthetic living materials in cancer biology
Nature Reviews Bioengineering (2023)
-
Fog robotics-based intelligence transportation system using line-of-sight intelligent transportation
Multimedia Tools and Applications (2023)