Abstract
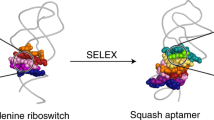
Fluorogenic RNA aptamers are used to genetically encode fluorescent RNA and to construct RNA-based metabolite sensors. Unlike naturally occurring aptamers that efficiently fold and undergo metabolite-induced conformational changes, fluorogenic aptamers can exhibit poor folding, which limits their cellular fluorescence. To overcome this, we evolved a naturally occurring well-folded adenine riboswitch into a fluorogenic aptamer. We generated a library of roughly 1015 adenine aptamer-like RNAs in which the adenine-binding pocket was randomized for both size and sequence, and selected Squash, which binds and activates the fluorescence of green fluorescent protein-like fluorophores. Squash exhibits markedly improved in-cell folding and highly efficient metabolite-dependent folding when fused to a S-adenosylmethionine (SAM)-binding aptamer. A Squash-based ratiometric sensor achieved quantitative SAM measurements, revealed cell-to-cell heterogeneity in SAM levels and revealed metabolic origins of SAM. These studies show that the efficient folding of naturally occurring aptamers can be exploited to engineer well-folded cell-compatible fluorogenic aptamers and devices.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information files. They are also available from the corresponding author on reasonable request. The following plasmids generated in this study will be available through Addgene: pAV-U6+27-Squash (ID 177913), pAV-5S-Squash (ID 177914), pAV-U6+27-Tornado-Squash (ID 177915), pAV-U6+27-Tornado-Squash-SAM sensor 4-2 (ID 177916), pAV-U6 + 27-Tornado-Squash-SAM sensor 5-1 (ID 177917), pAV-U6+27-Tornado-F30-Squash-Broccoli (ID 177918), pAV-U6+27-Tornado-F30-Broccoli-Squash-SAM sensor 4-2 (ID 177919) and pAV-U6+27-Tornado-F30-Broccoli-Squash-SAM sensor 5-1 (ID 177920). Source data are provided with this paper.
Code availability
The custom code used to analyze the sprouts and clips library is deposited in Bitbucket. It can be accessed through the following link:
https://bitbucket.org/jaffrey_lab_wcm/ncb_2021_dey_squash/src/master/.
Change history
21 January 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41589-022-00969-w
References
Sanford, L. & Palmer, A. Recent advances in development of genetically encoded fluorescent sensors. Methods Enzymol. 589, 1–49 (2017).
Lindenburg, L. & Merkx, M. Engineering genetically encoded FRET sensors. Sensors 14, 11691–11713 (2014).
Palmer, A. E., Qin, Y., Park, J. G. & McCombs, J. E. Design and application of genetically encoded biosensors. Trends Biotechnol. 29, 144–152 (2011).
Lechner, H., Ferruz, N. & Höcker, B. Strategies for designing non-natural enzymes and binders. Curr. Opin. Chem. Biol. 47, 67–76 (2018).
Paige, J. S., Nguyen-duc, T., Song, W. & Jaffrey, S. R. Fluorescence imaging of cellular metabolites with RNA. Science 335, 1194 (2012).
Kellenberger, C. A., Wilson, S. C., Sales-Lee, J. & Hammond, M. C. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 135, 4906–4909 (2013).
Sun, Z., Nguyen, T., McAuliffe, K. & You, M. Intracellular imaging with genetically encoded RNA-based molecular sensors. Nanomaterials 9, 233 (2019).
Kim, H. & Jaffrey, S. R. A fluorogenic RNA-based sensor activated by metabolite-induced RNA dimerization. Cell Chem. Biol. 26, 1725–1731 (2019).
Li, X. et al. Imaging intracellular S-adenosyl methionine dynamics in live mammalian cells with a genetically encoded red fluorescent RNA-based sensor. J. Am. Chem. Soc. 142, 14117–14124 (2020).
Ortega, A. D. et al. A synthetic RNA-based biosensor for fructose-1,6-bisphosphate that reports glycolytic flux. Cell Chem. Biol. https://doi.org/10.1016/j.chembiol.2021.04.006 (2021).
Litke, J. L. & Jaffrey, S. R. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 37, 667–675 (2019).
Frieda, K. L. & Block, S. M. Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338, 397–400 (2012).
Mandal, M. & Breaker, R. R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 11, 29–35 (2004).
Serganov, A. et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 11, 1729–1741 (2004).
Dalgarno, P. A. et al. Single-molecule chemical denaturation of riboswitches. Nucleic Acids Res. 41, 4253–4265 (2013).
Lemay, J. F., Penedo, J. C., Tremblay, R., Lilley, D. M. J. & Lafontaine, D. A. Folding of the adenine riboswitch. Chem. Biol. 13, 857–868 (2006).
Grubbs, R. D. Intracellular magnesium and magnesium buffering. BioMetals 15, 251–259 (2002).
Romani, A. M. P. Magnesium homeostasis in mammalian cells. in Metallomics and the Cell (ed. Banci, L.) 69–118 (Springer, 2013).
Tyrrell, J., Mcginnis, J. L., Weeks, K. M. & Pielak, G. J. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry 52, 8777–8785 (2013).
Paige, J. S., Wu, K. Y. & Jaffrey, S. R. RNA mimics of green fluorescent protein. Science 333, 642–646 (2011).
Filonov, G. S., Moon, J. D., Svensen, N. & Jaffrey, S. R. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 136, 16299–16308 (2014).
Song, W., Strack, R. L., Svensen, N. & Jaffrey, S. R. Plug-and-play fluorophores extend the spectral properties of spinach. J. Am. Chem. Soc. 136, 1198–1201 (2014).
Song, W. et al. Imaging RNA polymerase III transcription using a photostable RNA – fluorophore complex. Nat. Chem. Biol. 13, 1187–1194 (2017).
Steinmetzger, C., Palanisamy, N., Gore, K. R. & Höbartner, C. A multicolor large stokes shift fluorogen-activating RNA aptamer with cationic chromophores. Chem. Eur. J. 25, 1931–1935 (2019).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Dixon, N. et al. Reengineering orthogonally selective riboswitches. Proc. Natl Acad. Sci. USA 107, 2830–2835 (2010).
Porter, E. B., Polaski, J. T., Morck, M. M. & Batey, R. T. Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors. Nat. Chem. Biol. 13, 295–301 (2017).
Hall, B. et al. Design, synthesis, and amplification of DNA pools for in vitro selection. Curr. Protoc. Mol. Biol. 24, 24.2.1–24.2.27 (2009).
Bartel, D. P., Zapp, M. L., Green, M. R. & Szostak, J. W. HIV-1 rev regulation involves recognition of non-watson-crick base pairs in viral RNA. Cell 67, 529–536 (1991).
Sunbul, M. et al. Super-resolution RNA imaging using a rhodamine-binding aptamer with fast exchange kinetics. Nat. Biotechnol. 39, 686–690 (2021).
Truong, L. & Ferré-D’Amaré, A. R. From fluorescent proteins to fluorogenic RNAs: tools for imaging cellular macromolecules. Protein Sci. 28, 1374–1386 (2019).
Li, X., Kim, H., Litke, J. L., Wu, J. & Jaffrey, S. R. Fluorophore-promoted RNA folding and photostability enables imaging of single broccoli-tagged mRNAs in live mammalian cells. Angew. Chemie Int. Edn 59, 4511–4518 (2019).
Strack, R. L., Disney, M. D. & Jaffrey, S. R. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat–containing RNA. Nat. Methods 10, 1219–1224 (2013).
Shu, D., Khisamutdinov, E. F., Zhang, L. & Guo, P. Programmable folding of fusion RNA in vivo and in vitro driven by pRNA 3WJ motif of phi29 DNA packaging motor. Nucleic Acids Res. 42, e10 (2014).
Filonov, G. S. et al. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem. Biol. 22, 649–660 (2015).
Su, X., Wellen, K. E. & Rabinowitz, J. D. Metabolic control of methylation and acetylation. Curr. Opin. Chem. Biol. 30, 52–60 (2016).
Moon, J. D. et al. Naturally occurring three-way junctions can be repurposed as genetically encoded RNA-based sensors. Cell Chem. Biol. 28, S2451-9456(21)00215-4 (2021).
Lu, C. et al. Crystal structures of the SAM-III/SMK riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat. Struct. Mol. Biol. 15, 1076–1083 (2008).
Wang, J. et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science 325, 435–439 (2009).
Shyh-Chang, N. et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226 (2013).
Sanderson, S. M., Gao, X., Dai, Z. & Locasale, J. W. Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat. Rev. Cancer 19, 625–637 (2019).
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Chambers, I. et al. Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 (2007).
Filipczyk, A. et al. Biallelic expression of nanog protein in mouse embryonic stem cells. Cell Stem Cell 13, 12–13 (2013).
Lemay, J. F. et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 7, e1001278 (2011).
Wu, R. et al. Genetically encoded ratiometric RNA-based sensors for quantitative imaging of small molecules in living cells. Angew. Chemie Int. Edn 58, 18271–18275 (2019).
Sunbul, M. & Jäschke, A. SRB-2: a promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Res. 46, e110 (2018).
Hao, X. et al. Immunoassay of S-adenosylmethionine and S-adenosylhomocysteine: the methylation index as a biomarker for disease and health status. BMC Res. Notes 9, 498 (2016).
Han, K. Y., Leslie, B. J., Fei, J., Zhang, J. & Ha, T. Understanding the photophysics of the Spinach-DFHBI RNA aptamer-fluorogen complex to improve live-cell RNA imaging. J. Am. Chem. Soc. 135, 19033–19038 (2013).
Endoh, T. & Sugimoto, N. Selection of RNAs for constructing ‘Lighting-UP’ biomolecular switches in response to specific small molecules. PLoS One 8, e60222 (2013).
Endoh, T. & Sugimoto, N. Signaling aptamer optimization through selection using RNA-capturing microsphere particles. Anal. Chem. 92, 7955–7963 (2020).
Paul, C. P. et al. Localized expression of small RNA inhibitors in human cells. Mol. Ther. 7, 237–247 (2003).
Carey, B. W., Finley, L. W. S., Cross, J. R., Allis, C. D. & Thompson, C. B. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 (2015).
Mentch, S. J. et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 22, 861–873 (2015).
Acknowledgements
We thank members of the Jaffrey and Finley laboratories for helpful comments and suggestions. We thank S. Daniska and J. DeLuca from the Oligo Synthesis Facility, Keck Biotechnology Resource Laboratory at Yale University for their help in establishing the protocol for the sprouts and clips library generation. We thank R. Rao for help with analysis of some mutant RNAs during initial developments of Squash. We also thank J. McCormick at the Flow Cytometry Core facility at Weill Cornell Medical College for help with FACS. This work was supported by NIH grant nos. R01NS064516 and R35NS111631, the American Diabetes Association Pathway to Stop Diabetes grant no. 1-18-VSN-02, the Weill Cornell Medicine Daedalus Fund for Innovation, and DoD ALSRP Research Grant AL1301111 to S.R.J., Starr Cancer Consortium grant no. I12-0051 to S.R.J. and L.W.S.F., NIH grant nos. F32GM120987 and T32 CA062948 to A.O.O.-G.
Author information
Authors and Affiliations
Contributions
S.R.J. and S.K.D. conceived and designed the experiments. S.K.D. carried out experiments and analyzed data. G.S.F. and A.O.O.-G. contributed on analysis of the phosphoramidite coupling conditions for sprouts/clips library. G.S.F. also contributed toward theoretical analysis of sprouts/clips library. B.T.J. and L.W.S.F. contributed toward culturing mES cells, synthesis of custom media, and design of metabolism experiments. S.R.J. and S.K.D. wrote the paper with help from all the authors.
Corresponding author
Ethics declarations
Competing interests
S.R.J. is the cofounder of Lucerna Technologies and has equity in this company. Lucerna has licensed technology related to Spinach, Broccoli and other RNA–fluorophore complexes. S.R.J. is a founder of Chimerna Therapeutics and has equity in this company.
Peer review
Peer review information
Nature Chemical Biology thanks Yamuna Krishnan and other anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of the fluorescence properties of different intermediates during development of Squash.
a, Fluorescence activation of DFHO by Squash and its precursors. Fluorescence was measured with 200 nM dye and 10 µM RNA (ex: 495 nm, em: 562 nm, except for Corn; ex: 505 nm, em: 545 nm). Data represent mean values ± s.d. for n = 3 independent experiments. Squash showed >2-fold activation of DFHO compared to Corn. b, Fluorescence of initial hits after Round 7 of SELEX. In vitro transcribed RNAs from each hit (1 µM final) were mixed with DFHBI-1T (10 µM final) and the fluorescence was measured (ex: 452 nm, em: 503 nm, except for Broccoli; ex: 472 nm, em: 507 nm). Data represent mean values ± s.d. for n = 3 independent experiments. c,d, Fluorescence measurements of the hits with DFHBI-1T after first (c) and second (d) round of directed evolution, measured as in panel b. Data represent mean values ± s.d. for n = 3 independent experiments. e, Fluorescence measurements of hits using DFHO after the second round of directed evolution measured as in panel a. The fluorescence of the hits were normalized against Corn. Data represent mean values ± s.d. for n = 3 independent experiments. f, DE2-6 has one bulged nucleotide (U51) and one G•U pair based on mFold (Supplementary Fig. 3d). Elimination of the bulge, the G•U pair, or both did not improve fluorescence of DE2-6. Fluorescence was measured as in a. Data represent mean values ± s.d. for n = 3 independent experiments. g, Mutation (G30C) of the kissing loop of DE2-6 resulted in loss of fluorescence which was recovered by a complementary mutation (C57G) in the opposite loop of the predicted kissing loop. Similar results were also observed for the other kissing loop basepair. Two mutations (U29C, U58G) created an extra G•C basepair in the kissing loop compared to add A-aptamer and resulted in ~20% increase in fluorescence of DE2-6. Further improvement of the kissing loop interaction cannot be achieved by further mutations (A28G + U59C in Squash). Data represent mean values ± s.d. for n = 3 independent experiments. h, Although Broccoli–BI and Squash-DFHO (20 µM RNA, 2 µM dye) excitation spectra overlap, their emission maxima show ~57 nm separation. Structure of BI is shown in the inset.
Extended Data Fig. 2 Photophysical and biophysical characterization of Squash.
a, Absorbance spectra (50 µM RNA, 5 µM DFHBI-1T) of DFHBI-1T alone and in complex with Squash demonstrates a smaller red shift (Abs max: 450 nm) compared to Spinach (Abs max: 470 nm)20 and Broccoli (Abs max: 469 nm)21. Excess RNA was used to ensure binding of nearly all fluorophore. b, DFHO shows a red-shifted absorbance spectrum upon binding Squash (50 µM RNA, 5 µM DFHO). c, Both Squash and Broccoli shows similar emission maxima. Squash also showed higher fluorescence (10 µM RNA, 1 µM DFHBI-1T, ex. 452 nm for Squash and 472 nm for Broccoli) intensity with DFHBI-1T compared to DE2-6. d, Fluorescence spectra (10 µM RNA, 1 µM DFHO, ex. 495 nm for Squash and 505 nm for Corn) showed a red-shifted Squash emission maxima (562 nm) compared to Corn (545 nm), consistent with a different mode of interaction between DFHO and Squash compared to Corn. Squash also showed more than two fold higher fluorescence intensity with DFHO compared to Corn. e, Kd was measured by titration of 50 nM RNA with DFHBI-1T and then fitting the data using a one-site saturation model. Data represent mean values ± s.d. for n = 3 independent experiments. f, Binding kinetics were performed as reported previously50. The fluorescence signal trace was fitted with a monoexponential curve to extract kobs. kobs was plotted as a function of total RNA (50 nM RNA) and fluorophore concentration. kon and koff were extracted as the slope and intercept, respectively. Squash koff (0.014 ± 0.008 s-1) is very similar to that of Corn (0.018 ± 0.002 s-1). kon for Squash-DFHO (162300 ± 8100 M-1 s-1) is seven-times higher than Corn-DFHO (23000 ± 3000 M-1 s-1) which could be due to the high folding of Squash. g,h, Kd measurements for Broccoli and Squash binding to BI (g) and DFHO (h) were performed as in panel e, using 50 nM RNA and then fitting the data using a one-site saturation model. Data represent mean values ± s.d. for n = 3 independent experiments.
Extended Data Fig. 3 Comparison of in cell photostability of Squash with other fluorogenic aptamers.
a, Comparison of in cell photostability of Squash-DFHBI-1T pair with Broccoli-BI. Compared to DFHBI-1T, BI shows improved photostability when bound to Broccoli33. We compared the photostability of Squash-DFHBI-1T with Broccoli-BI. Aptamers were expressed as circular RNAs11 and aptamer-expressing cells were incubated with dyes (10 µM). Continuous images (100 ms per frame) were taken for 10 s total while the cells were continuously illuminated. We used the highest available light power in the microscope since previously lower light levels were used and Broccoli-BI photobleaching was not readily detected33. Shown are every other frame starting from 0 ms to 1000 ms. Scale bar, 20 µm. b, Comparison of in cell photostability of Squash-DFHO with Corn-DFHO. Corn aptamer exhibits high photostability with DFHO compared to Spinach or Broccoli with DFHBI-1T23. We compared the photostability of Squash-DFHO with tCorn (Corn in the tRNA scaffold)-DFHO. The images were taken in as in panel a except the exposure time was 500 ms per frame. Shown are frame between 0 and 2500 ms. Scale bar, 20 µm. c, Quantification of in cell photostability for green emitting aptamers. Cellular mean fluorescence intensity was calculated and normalized to maximum intensity at 0 s. The normalized cellular fluorescence intensities were plotted against time. Both Broccoli-BI and Squash-DFHBI-1T showed a similar initial rate of drop in fluorescence, which reflects the rate of light-induced photoisomerization of the fluorophore to the low-fluorescence trans-isomer33. The higher plateau associated with Squash-DFHBI-1T likely reflects a higher on-rate for fluorophore binding since the plateau reflects the balance between light-induced isomerization/trans-fluorophore dissociation and cis-fluorophore rebinding33. d, Quantification of in cell photostability for yellow-orange emitting aptamers. Assays were performed as in c. For comparison, we used mVenus, a fluorescent protein which has similar fluorescence emission properties as Corn-DFHO.
Extended Data Fig. 4 Squash exhibits higher cellular fluorescence than Corn when expressed as a circular RNA.
a, Comparison of in-cell fluorescence intensity of circular Squash or circular tCorn (tRNA-scaffolded Corn). HEK293T cells expressing 5S-control (no aptamer), Tornado-tCorn or Tornado-Squash, plasmids were imaged using 10 µM DFHO and the same microscope settings. Cells expressing circular Squash exhibited much brighter cellular fluorescence than cells expressing circular tCorn. Circular Squash showed mostly nucleus-excluded signal. Scale bar, 20 μm. b, Comparison of cellular brightness of Squash and Corn using flow cytometry of HEK293T cells prepared as in a. Cells were analyzed in the yellow (ex 488 nm, em 545 ± 17.5 nm) fluorescence channel. An auxiliary far-red channel (ex 635 nm; em 780 ± 30) was used to measure cellular auto-fluorescence. Cells expressing circular Squash exhibited substantially more fluorescence. c, To determine if the higher fluorescence of circular Squash in a and b was due to higher expression, 10 µg total RNA was isolated from HEK293T cells and analyzed by gel staining with DFHO and SYBR Gold as described previously36. Bands corresponding to circular Squash and circular tCorn in SYBR Gold staining were identified by comparing them with the 5S-control lane and marked by white stars. Quantification of the band intensities indicates slightly higher expression of circular tCorn. Note, unlike Squash, two tCorn molecules are required to bind a single DFHO molecule. The higher fluorescence of circular Squash-expressing cells compared to tCorn is probably due to higher quantum yield and improved folding of Squash. Additionally, the DFHO fluorescence intensity of the band corresponding to circular Squash is much higher than that of the circular tCorn band, suggesting that Squash also folds better in the gel than Corn.
Extended Data Fig. 5 Development and characterization of Squash-SAM sensors.
a, Squash was fused with the SAM-binding SAM-III aptamer through a sprouts and clips randomized transducer (NnNnNnNn for each strand). 2 µM of the RNA pool after each SELEX round was mixed with 10 µM DFHO and fluorescence was measured in the absence and presence of 0.1 mM SAM at 37 ˚C. b, For each library member, 1 µM of the in vitro transcribed RNA was incubated with DFHO in the absence or presence of 0.1 mM SAM (37 ˚C, 10 min) and put into separate tubes and imaged immediately (ex: 530 ± 14 nm, em = 605 ± 25 nm). Squash-SAM sensors 4-2 and 5-1 are highlighted. c,f, Squash-SAM sensor 4-2 (c) and 5-1 (f) are activated by SAM and not related molecules. 1 µM RNA was mixed with 10 µM DFHO and indicated molecules. Data represent mean values ± s.d. for n = 3 independent experiments. d,g, Measurement of Kd of Squash-SAM sensor 4-2 (d) and 5-1 (g) with DFHO were measured as in Extended Data Fig. 2e. The Kds are slightly weaker than that of Squash with DFHO (see Fig. 2d). Data represent mean values ± s.d. for n = 3 independent experiments. e,h, Measurement of Kd of Squash-SAM sensor 4-2 (e) and 5-1 (h) with SAM. Sensor RNA (1 µM) was titrated with SAM in presence of 10 µM DFHO (37 ˚C) and the resulting data was fitted using one-site saturation model. The Kd for the SAM-III riboswitch is ~1 µM. Data represent mean values ± s.d. for n = 3 independent experiments. i, Broccoli-BI fluorescence (1 µM Broccoli, 10 µM BI) is not affected by SAM or related molecules making it an appropriate normalizer for ratiometric sensor. Data represent mean values ± s.d. for n = 3 independent experiments.
Extended Data Fig. 6 Detection of intracellular SAM levels using Squash-based SAM sensors.
a, Detection of intracellular SAM levels using the Squash-SAM sensor 4-2 and 5-1. HEK293T cells were transfected with the Tornado plasmids expressing the sensors or the constitutively fluorescent Squash aptamer (no SAM dependent fluorescence enhancement) as circular RNAs. Cells expressing the circular SAM biosensors or circular Squash were incubated with 10 µM DFHO and imaged every 10 min after treatment with 30 mM cycloleucine (an inhibitor of the major SAM biosynthesis enzyme, MAT2A) for 90 min total. Following this, cycloleucine was removed by replacing the imaging media with cycloleucine-free media. Images were taken again at 10 min intervals for 90 min. Images are shown at specific intervals along with results for representative cells. Cells expressing the circular sensors showed drop in the cellular fluorescence during cycloleucine treatment and increase in cellular fluorescence during cycloleucine withdrawal. However, cell expressing circular Squash did not show any change in cellular fluorescence during this treatment. Scale bar, 20 μm. b,c, Quantification of live-cell SAM levels based on Squash-SAM sensors 4-2 (b) and 5-1 (c) respectively. Cellular mean fluorescence intensity was calculated for each sensor and normalized to maximum intensity at time point 0. The normalized values of mean cellular fluorescence intensity were plotted against time at 10 min intervals for six different cells. Each cell is represented using a different color. For each sensor, SAM decay was observed following addition of cycloleucine and SAM recovery is seen after withdrawal of cycloleucine. Interestingly for both the sensors, the SAM decay profile for each individual cell is very similar while the SAM recovery profiles are distinctly different.
Extended Data Fig. 7 Broccoli-BI and Squash-DFHO can be used as an orthogonal pair of fluorogenic aptamers for two color imaging.
a, Lack of correlation of eqFP670 and Squash fluorescence expressed from the same plasmid (U6+27 promoter for Squash and PGK for eqFP670). Many HEK293T cells have high Squash fluorescence and low eqFP670 fluorescence (white arrows). Some cells have correlated fluorescence (gray arrows). Flow cytometry also revealed poor correlation. Scale bar, 20 μm. b, Schematic representation of the circular F30-Broccoli-Squash RNA used as a constitutive control for the ratiometric sensor (Fig. 4f). The RNA was generated by fusing Squash and Broccoli to each arm of an F30 scaffold. c, High correlation between Squash and Broccoli fluorescence when expressed within a single RNA. The F30-Broccoli-Squash construct (panel b) was expressed as a circular RNA in HEK293T cells and imaged in both green and orange channels after incubation with 10 µM BI and 10 µM DFHO. Excellent correlation was observed for each cell (see Fig. 4f, bottom panel for O/G ratio). This was also confirmed by flow cytometry. d, Circular Broccoli-expressing cells showed negligible bleedthrough in the orange channel and circular Squash-expressing cells showed no bleedthrough in the green channel. Circular F30-Broccoli-Squash shows bright signal in both channels. e, To determine if DFHO can compete with BI’s ability to bind Broccoli, we imaged HEK293T cells expressing circular Broccoli (10 µM BI) and compared green fluorescence before and after addition of DFHO (10 µM, 1 hr). The green fluorescence signal was not substantially affected. Similarly, BI does not affect Squash-DFHO fluorescence when circular Squash is expressed in HEK293T cells. f, To test the possibility of FRET in the ratiometric sensor, we expressed ratiometric Squash-SAM sensor 5-1 in HEK293T cells as a circle. After incubating the cells with 10 µM BI, we took an image in the green channel. Then we added DFHO (10 µM, 1 hr incubation). No decrease in the Broccoli-BI (possible donor) green fluorescence was seen upon formation of the Squash-DFHO pair (possible acceptor), consistent with lack of FRET.
Extended Data Fig. 8 Quantification of cellular SAM concentration using ratiometric sensors.
a, Standard curve measuring FRET signal (ex: 485 nm, em: 665 nm) in a biochemical assay to determine SAM concentration (see Methods). Data represent mean values ± s.d. for n = 3 independent experiments. b, The average cellular concentration of SAM was measured from 1.2 ×106 cells utilizing the standard curve in panel a. To calculate cellular volume, we assumed HEK293T cells are 17 µm-diameter spheres. We quantified the average SAM concentration at different time points during cycloleucine treatment and withdrawal. SAM concentrations were not affected by sensor expression. Data represent mean values ± s.d. for n = 3 independent experiments. c,d, Correlation between the ratiometric signal and biochemical SAM measurements for Squash-SAM sensors 5-1 (c) and 4-2 (d). To convert the sensor ratiometric signal into SAM concentration, we calculated the average ratiometric signal for HEK293T cells during cycloleucine treatment (n = 6 cells, from Fig. 4g). Using the biochemical assay we determined the average SAM concentration at the same time points, allowing extrapolation of intracellular SAM concentrations from the O/G ratio. e, We measured the average O/G fluorescence ratio in 30 cells from three independent measurements for each indicated cell type and culture condition. HEK293T cells and mES cells cultured in +2i media showed a narrow distribution while HCT116 and mES cells cultured in -2i showed a wide range of O/G ratios. Sensors used for each cell type are indicated. Line indicates median, box shows the interquartile range, and whiskers are the minimum and maximum values. f, The average O/G fluorescence ratio for each cell in panel e was converted into average cellular SAM concentration (based on panel c and d) and plotted for each condition. For HEK293T cells, the distribution of O/G fluorescence ratios was different for the two Squash-SAM sensors; however, when these O/G ratios are converted to SAM concentrations based on the specific O/G-to-SAM correlation for each sensor, the cellular SAM concentrations were similar regardless of the sensor. Line indicates median, box shows the interquartile range, and whiskers are the minimum and maximum values.
Extended Data Fig. 9 Effect of cycloleucine treatment and different amino acids depletion on SAM level in mESCs.
a, We expressed both Squash-SAM sensors 4-2 and 5-1 for ratiometric SAM imaging in mESCs. Unlike HEK293T cells, the SAM-dependent Squash signal (DFHO channel) was too dim for the 5-1 sensor. However, Squash-SAM sensor 4-2 gave much brighter cellular fluorescence signal in the DFHO channel. Thus, despite having lower dynamic range than Squash-SAM sensor 5-1, we used the Squash-SAM sensor 4-2 for ratiometric imaging of SAM in mESCs. Image acquisition time was 100 ms for BI channel and 500 ms for DFHO channel. Scale bar, 20 μm. b, To test whether MAT2A is the major SAM biosynthetic enzyme in mESCs, we expressed Squash-SAM sensor 4-2 and treated the cells with cycloleucine (30 mM final), an MAT2A inhibitor. A rapid drop of cellular SAM levels was observed in 30 min for mESCs cultured in both +2i and −2i media. Note: mESCs cultured in −2i media exhibit a large flattened morphology, which accounts for their increased size relative to mESCs cultured in +2i. Scale bar, 20 μm. c,d, The drop in SAM levels due to amino acid depletion in mESCs is reversible in cells cultured in either +2i (c) or −2i (d). In Fig. 5a, we showed that depletion of methionine leads to a drop in SAM in mESCs. To see if reintroduction of the amino acids in the media leads to rise in SAM level, we added the amino acids back to the media. Images are shown at indicated time points. For most mESCs cultured in both +2i and −2i conditions, we saw that the SAM levels go back to the pre-treatment condition, indicating the reversible nature and lack of cytotoxicity of this process. Scale bar, 20 μm.
Extended Data Fig. 10 Effect of different amino acids depletion and cycloleucine treatment on SAM level in HCT116 cells.
a, Both Squash-SAM sensors 4-2 and 5-1 produced signal that is substantially higher than the background in both the channels. We used 5-1 because it has higher dynamic range. Scale bar, 20 μm. b, Threonine depletion does not change SAM levels in mESCs (see Fig. 5a). To test if this also holds in HCT116 cells, we expressed Squash-SAM sensor 5-1 in HCT116 cells and monitored SAM level after threonine depletion. Most cells did not show any notable change in SAM levels, consistent with a lack of functional threonine dehydrogenase enzyme in these cells. c, Although threonine depletion does not change SAM levels (panel b), threonine could be important in methionine-depleted cells. To test this, we expressed Squash-SAM sensor 5-1 in HCT116 cells and monitored SAM levels for 240 min after depleting both threonine and methionine. Threonine depletion did not exacerbate the drop in SAM levels, suggesting that threonine does not have a major role in SAM levels in these cells. d, Circular Squash-SAM sensor 5-1 expressed in HCT116 cells showed marked drop in SAM after cycloleucine (30 mM final) treatment, indicating MAT2A is required for SAM biosynthesis in these cells. A rapid drop in SAM levels was observed in 30 min for most cells. The cells indicated by white arrows (in c and d) start with a very high level of SAM and show slower drop in SAM level over time compared to other cells. e, HCT116 cells have three different cell populations based on baseline SAM levels (see Fig. 5d,e). We quantified the average cellular ratio of O/G fluorescence for these cells at 15 min intervals after cycloleucine (30 mM final) treatment. All three populations showed slightly faster drop in SAM levels with cycloleucine treatment compared to depletion of methionine and threonine together.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1–5 and Note.
Source data
Source Data Extended Data Fig. 4
Unprocessed gel images for Extended Data Fig. 4c.
Rights and permissions
About this article
Cite this article
Dey, S.K., Filonov, G.S., Olarerin-George, A.O. et al. Repurposing an adenine riboswitch into a fluorogenic imaging and sensing tag. Nat Chem Biol 18, 180–190 (2022). https://doi.org/10.1038/s41589-021-00925-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00925-0
This article is cited by
-
Quiescence enables unrestricted cell fate in naive embryonic stem cells
Nature Communications (2024)
-
Fluorogenic CRISPR for genomic DNA imaging
Nature Communications (2024)
-
p16High senescence restricts cellular plasticity during somatic cell reprogramming
Nature Cell Biology (2023)
-
Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture
Nature Communications (2023)
-
Visualizing orthogonal RNAs simultaneously in live mammalian cells by fluorescence lifetime imaging microscopy (FLIM)
Nature Communications (2023)