Abstract
Emerging evidence suggests that host glycans influence severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Here, we reveal that the receptor-binding domain (RBD) of the spike (S) protein on SARS-CoV-2 recognizes oligosaccharides containing sialic acid (Sia), with preference for monosialylated gangliosides. Gangliosides embedded within an artificial membrane also bind to the RBD. The monomeric affinities (Kd = 100–200 μM) of gangliosides for the RBD are similar to another negatively charged glycan ligand of the RBD proposed as a viral co-receptor, heparan sulfate (HS) dp2–dp6 oligosaccharides. RBD binding and infection of SARS-CoV-2 pseudotyped lentivirus to angiotensin-converting enzyme 2 (ACE2)-expressing cells is decreased following depletion of cell surface Sia levels using three approaches: sialyltransferase (ST) inhibition, genetic knockout of Sia biosynthesis, or neuraminidase treatment. These effects on RBD binding and both pseudotyped and authentic SARS-CoV-2 viral entry are recapitulated with pharmacological or genetic disruption of glycolipid biosynthesis. Together, these results suggest that sialylated glycans, specifically glycolipids, facilitate viral entry of SARS-CoV-2.

Similar content being viewed by others
Main
Many viruses exploit carbohydrates, also known as glycans, attached to protein and lipid carriers on host epithelial cells for viral entry1. Sialoglycans are acidic Sia-containing glycans (for example, gangliosides, mucin-type O-glycans and complex N-glycans) that are densely displayed on the surface of mammalian cells2. They act as co-receptors for a wide variety of viruses, including orthomyxoviruses, paramyxoviruses, picornaviruses, reoviruses, polyomaviruses, adenoviruses, calicivirus and parvoviruses3. SARS-CoV-2, which is responsible for the global outbreak of coronavirus disease 2019 (COVID-19) and a member of the coronavirus family, is believed to rely on a combination of ACE2 and glycans to bind and infect the lungs as well as other tissues and organs4,5,6. Human coronaviruses generally rely on glycans to assist in cell entry7. For example, Middle East respiratory syndrome coronavirus (MERS-CoV) binds sialoglycans to facilitate cellular entry8, and the human betacoronaviruses OC43 and HKU1 engage sialoglycans with 9-O-acetylated Sia as key receptors9, while SARS-CoV-1 and CoV-NL63 exploit acidic HS polysaccharides10,11.
There is emerging evidence that acidic glycans serve as co-receptors for SARS-CoV-2. Electrospray ionization mass spectrometry (ESI-MS) analysis revealed that binding of oligosaccharide fragments of heparin, a highly sulfated form of HS, to the RBD of the transmembrane S glycoproteins of SARS-CoV-2 inhibits ACE2 binding12. Consistent with this finding, free HS inhibited SARS-CoV-2 infection of Vero cells13. HS was reported to enhance the affinity of the SARS-CoV-2 RBD for ACE2, suggestive of HS acting as a more classical co-receptor14. Notably, destroying cellular HS enzymatically with heparanase, or competing with unfractionated heparin, substantially reduced SARS-CoV-2 infection14. It was speculated early on that SARS-CoV-2 may also exploit sialoglycans on cells15, with some evidence suggesting that Sia can bind SARS-CoV-2. Results of biolayer interferometry showed that Sia-conjugated gold nanoparticles exhibit high avidity for the SARS-CoV-2 S1 protein, which contains both the N-terminal domain and RBD of the S protein16. Efforts to quantify these interactions on model sialylated species (for example, 3′-sialyllactose), however, were challenging and revealed weak binding (Kd millimolar)16. Given this weak binding, it is not surprising that conflicting reports exist for the binding of the RBD to sialylated species by traditional glycan microarray screening6,17,18. Beyond acidic glycans, recent work suggests that the RBD mimics a galectin scaffold and can bind blood group A antigens19 and terminal N-acetylglucosamine moieties (for example, GlcNAcα1-3GalNAcβ)17. Moreover, it was recently reported that sialylated N- and O-glycans on ACE2 may play a role in S protein cell binding20,21.
To further examine the role of glycans in SARS-CoV-2 infection, we analyzed the binding of a library of glycans to SARS-CoV-2 RBD. Using catch-and-release ESI-MS (CaR-ESI-MS)22,23, a label-free and semi-quantitative screening method capable of detecting weak, yet biologically relevant, interactions within a complex mixture, we discovered that several classes of sialoglycans bind to the RBD, with ganglioside oligosaccharides being the top hits. Cell-based studies revealed that RBD binding and SARS-CoV-2 pseudotyped virus entry in ACE2-expressing cells is decreased following depletion of Sia levels on cells pharmacologically, genetically or enzymatically. Blocking glycolipid biosynthesis produced similar decreases in RBD binding and viral infection of both pseudotyped and native virus, pointing to RBD–glycolipid interactions as being important for SARS-CoV-2 infection of cells.
Results
CaR-ESI-MS screening of glycan libraries
A total of 139 glycans (Extended Data Fig. 1), consisting mostly of mammalian glycans (1–132)24,25,26,27 but also several non-mammalian rhamnose-containing oligosaccharides as negative controls (133–139)28, were used for screening against the SARS-CoV-2 RBD and S protein using the CaR-ESI-MS assay. To ensure that all species were of different molecular weights (MWs), the library was divided into 16 sublibraries of glycans (libraries A–P; Supplementary Table 1 and Supplementary Fig. 1). In all cases, a reference protein (Pref) was added to control for non-specific glycan–RBD interactions during ESI29.
Numerous glycan ligands were released from the RBD in CaR-ESI-MS performed at both at 25 °C (Extended Data Fig. 2) and 37 °C (Fig. 1a). The low relative abundances of released ligands are indicative of generally low affinities. No binding was detected for the non-mammalian rhamnose-containing glycans (133–139) or any detectable glycans released from Pref. These results indicate that the low-abundance ligands released from the RBD are the result of specific interactions and not false positives. At 37 °C, the pentasaccharide (69) of the ganglioside GM1 was the preferred ligand. Other gangliosides were recognized, with a preference for monosialylated (69–72) over di- and trisialylated gangliosides. Acidic human milk oligosaccharides (HMOs) were consistently observed at high relative abundances, and some of the ABH blood group antigens (in particular A and H types 2, 3 and 4) exhibited moderate CaR-ESI-MS signal. Interestingly, chondroitin sulfate (33 and 34) and heparosan-derived oligosaccharides containing GlcA (35 and 36), which are acidic glycans, showed little to no binding to RBD. These results, when considered with the preference for monosialylated over di- and trisialylated ganglioside oligosaccharides, argue against electrostatics being the dominant force underpinning RBD–ligand binding. Overall, the screening results obtained at 25 °C are similar to those obtained at 37 °C in terms of the glycans recognized, with only minor changes in the relative abundances for some ligands (Extended Data Fig. 2). HS glycans (39–41) were not included in CaR-ESI-MS screening due to difficulties in releasing them from RBD at the energies used to screen the other glycans; the higher energies required for HS release promote fragmentation of the other ligands. The greater stability of the HS-bound RBD does not imply higher affinity than the other ligands, as trends in gas-phase kinetic stability of protein–ligand complexes do not necessarily reflect trends in Kd30. Notably, the relative abundances of the HS shown in Fig. 1a were estimated from their affinities, measured by ESI-MS, relative to those of the highest affinity ligands identified by screening. On the basis of this analysis, HS 41 was predicted to have a CaR-ESI-MS response slightly lower than that of 69.
a,b, Normalized abundances of released glycans from the SARS-CoV-2 RBD and S protein by CaR-ESI-MS at 37 °C. Summary of the charge-normalized (relative to 69) abundances (Abrel) of released ligands (L) measured by CaR-ESI-MS screening of libraries A–O against SARS-CoV-2 RBD (a) and S protein (b). Measurements were performed in negative ion mode with an ultrahigh mass range (UHMR) Orbitrap mass spectrometer on aqueous ammonium acetate (100 mM, pH 6.9) solutions of RBD (10 μM) and glycan library (A–O) and library P (50 nM of each glycan) or S protein (280 μg ml–1) and glycan library (A–O) and library P (500 nM of each glycan). A high-energy C-trap dissociation (HCD) energy of 50 V (using nitrogen as collision gas) with an isolation window of m/z 3,000–4,000 was used for screening against RBD; an HCD energy of 300 V (using argon) with an isolation window of m/z 8,000–15,000 was used for screening against S protein. The different classes of oligosaccharides are distinguished by color: red, Lewis antigens (1–14); yellow, blood group A antigens (15–20); orange, blood group B antigens (21–26); green, blood group H antigens (27–32); blue, sulfated compounds (34–38) and HS oligosaccharides (39–41); light blue, antigen-related glycans (42–50); purple, globo (51–60); pink, ganglioside oligosaccharides (61–76); white, HMOs and other glycans (77–128); light gray, Neu5Acα2-6-linked oligosaccharides (129–132); dark gray, rhamnose-containing compounds (133–139); NA, not applicable. An asterisk indicates that the relative abundances (in CaR-ESI-MS) were estimated from their relative (to 69) Kd. Data represent mean ± s.d.; n = 3 independent experiments. c, Affinities of glycans (15, 21, 27, 35, 36, 39–41, 69, 71, 76, 129 and 140) for RBD. Kd (μM) values were measured by ESI-MS using aqueous ammonium acetate (100 mM, pH 6.9) solutions containing RBD (5 μM) and each glycan (three different concentrations ranging from 30 to 150 μM) at 25 °C (white bars) and 37 °C (black bars); ND, not detected. Data represent mean ± s.d.; n = 3 independent experiments for each glycan concentration.
We next performed CaR-ESI-MS screening of the sublibraries against the S protein at 37 °C (Fig. 1b). The majority of ligands identified for the S protein were detected for the RBD and, notably, 69 was also the top hit for the S protein. It is also notable that the relative CaR-ESI-MS responses for neutral glycans, in particular for blood group antigens and structurally related glycans (for example, glycans 4 and 115) and xenoantigen α-Gal-Lewis X (50), were enhanced for the S protein.
Affinities of RBD toward select glycans
The direct ESI-MS binding assay, which relies on direct detection of intact complexes, was used to quantify glycan ligand affinities (Kd). The SARS-CoV-2 S protein possesses 26 glycosylation sites (22 N- and 4 O-glycosylation sites)31. This extensive degree of glycosylation, together with the appearance of multiple oligomeric states (monomer, dimer, trimer and hexamer) in ESI-MS analysis (Supplementary Fig. 2a), makes the application of direct ESI-MS analysis to the S protein challenging. Consequently, the RBD, which contains only two N-glycosylation sites (N331 and N343) and two O-glycosylation sites (T323 and S325), was selected for direct ESI-MS assay (Supplementary Fig. 2b,c)32,33,34,35. Quantifying interactions of ligands to individual glycoforms of the RBD by ESI-MS is challenging because of the unknown concentration of each glycoform and the potential for spectral overlap (ligand-bound and free RBD species). Therefore, determining the affinities of RBD for glycans required the elucidation of glycoforms and an estimation of their respective concentrations, which assumed that all glycoforms have uniform ESI-MS response factors. A total of 77 distinct RBD MWs were identified by ESI-MS (Supplementary Table 2 and Extended Data Fig. 3). According to reported glycomics studies, RBD produced from HEK293 cells has predominantly complex-type N-glycans and core 1 and 2 mucin-type O-glycans (Supplementary Table 3)32,33,34,35. Hydrophilic interaction-ultra high-performance liquid chromatography with fluorescence detection (HILIC-UHPLC-FLD)/ESI-MS analysis of the N-glycans released from the RBD sample used in this work identified 136 distinct structures with 74 distinct compositions (Supplementary Table 4 and Supplementary Fig. 3). Based on the reported O-glycans and results of the current N-glycan analysis, the glycan composition of each RBD species was putatively assigned. It was found that the major glycoforms possess two to eight Neu5Ac residues and are di- and trifucosylated (Supplementary Fig. 3).
The affinities of the top hits (gangliosides 69 and 71) and moderate binders (15, 21, 27, 76 and 129) identified by CaR-ESI-MS screening of the defined library, as well as the HS 39–41 for the RBD, were measured by ESI-MS at pH 6.9 and both 25 °C and 37 °C to estimate the binding enthalpy. Two non-binders (35 and 36) were also included as negative controls. The reported affinities are the average values measured for all the RBD species detected (Fig. 1c). It is notable that RBD glycosylation has a minimal effect on the measured Kd (Supplementary Fig. 4), suggesting that the glycosylation sites are remote from the glycan-binding site(s) and do not strongly influence the conformations of glycan-binding motifs within the RBD.
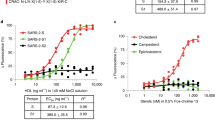
The measured affinities for the RBD are all ≥100 μM, with ganglioside 69 (GM1) having the highest affinity (160 ± 40 μM) at 25 °C. For the HS 39–41, the trend in affinities is 39 ≈ 40 > 41, with Kd values ranging from 200 μM to 400 μM. With the exception of 76 (which was undetectable at 37 °C), the temperature dependence of the measured Kd was relatively minor, with changes of less than 50%. Based on differences in affinities, the average association enthalpy change was –3.4 kcal mol–1, which is modest for protein–glycan interactions; however, the entropy change is favorable (with average entropy changes of 10–20 cal K–1 mol–1). The trend in affinities agrees with the trends in relative abundances measured by CaR-ESI-MS (Extended Data Fig. 4), establishing the reliability of CaR-ESI-MS for identifying glycan ligands and differentiating the high-affinity ligands from the low-affinity ligands and non-binders.
Tissue-derived N-glycan libraries
Direct evidence of binding between the S protein or RBD and host N-glycans is lacking. To assess the N-glycan-binding properties of RBD, libraries were prepared from human lung and intestinal tissues and screened against RBD by CaR-ESI-MS at 25 °C and 37 °C. At 37 °C, screening of the lung tissue-derived N-glycans detected ligands with 19 distinct MWs consisting of 13 mono- and 6 disialylated N-glycans, of which six were hybrid type and the remainder were complex type. The absence of any trisialylated N-glycan ligands may reflect their low abundances and low affinities compared to those of the mono- and disialylated N-glycans (Fig. 2a). Two of the hits detected by CaR-ESI-MS, m/z of 1,549.55 ([M – H]–, HexNAc3Hex3 Fuc1Neu5Ac1) and 1,889.65 ([M – H]–, HexNAc3Hex6Neu5Ac1), were not observed in HILIC-UHPLC-FLD analysis, suggesting low abundance yet high affinity (Fig. 2b). Ligands with 29 distinct MWs were detected from screening of the intestinal tissue N-glycans (Fig. 2c). The hits consisted of 14 mono-, 7 di- and 5 trisialylated N-glycans, of which 6 were hybrid type and the rest were complex type. Three of these hits (HexNAc3Hex3Neu5Ac1, HexNAc3Hex5Neu5Ac1 and HexNAc3Hex6Neu5Ac1) were not observed in the HILIC-FLD analysis, presumably due to their low abundances. Notably, no neutral N-glycan ligands were detected for either library. With the exception of the glycan HexNAc4Hex5Neu5Ac2, which was the most abundant glycan in the intestinal library (Fig. 2d), no N-glycans were released from Pref in any of the CaR-ESI-MS measurements, indicating an absence of non-specific binding to the remaining hits.
a,c, CaR-ESI-MS screening results obtained for aqueous ammonium acetate solutions (100 mM, pH 6.9, 37 °C) of RBD (10 μM), P dimer of the Saga strain (Pref, 4 μM) and the N-glycan library from lung (200 μg ml–1) (a) and intestinal (125 μg ml–1) (c) tissues. Ions with an m/z of 3,000–4,000 were subjected to HCD using a collision energy of 50 V. b,d, Chromatograms of 2-aminobenzamide (2-AB)-labeled N-glycans released from lung (b) and intestinal (d) tissues acquired using HILIC with fluorescence detection. The relative abundances of the Neu5Ac content of the N-glycans are indicated graphically. e, Heat maps of relative affinities of ligands identified by CaR-ESI-MS screening at 37 °C; ND, not detected.
On the basis of the relative abundances of released N-glycans measured by CaR-ESI-MS and their relative concentrations in each library (deduced from UHPLC analysis), the relative affinities of the N-glycan ligands were ranked (Fig. 2e). From the resulting heat maps, it can be seen that the RBD has a strong preference for monosialylated, monoantennary N-glycans, with the top hit having the composition HexNAc3Hex4Neu5Ac1. Based on the calculated glucose unit values from elution times of N-glycans (the HPLC elution position, expressed as glucose unit value, for each glycan is related to the number and the linkage types of its constituent monosaccharides), this ligand is sialylated on the α-(1-3)-arm36. To relate the CaR-ESI-MS results acquired for the defined and natural N-glycan libraries, we also performed quantitative binding measurements on glycan 140 (HexNAc3Hex4Neu5Ac1 with a Neu5Acα2-3 linkage), which has the same glycan composition as the top hit identified from the screening. The affinity for RBD at 25 °C and 37 °C was 460 ± 30 μM and 600 ± 30 μM, respectively (Fig. 1c).
CaR-ESI-MS screening of gangliosides
The CaR-ESI-MS screening revealed a preference for oligosaccharides derived from gangliosides, with GM2 and GM1 being the top hits. As gangliosides are glycolipids normally embedded within a lipid bilayer, we screened the RBD against a library of six gangliosides (GM1, GM2, GM3, GD1a, GD2 and GT1b, each nominally 1% of total lipid), presented together in a nanodisc (ND) composed of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)37. To positively identify the gangliosides bound to the RBD, ions within a 100-m/z window centered at m/z 3,540, which encompasses ions at charge state –9 of any RBD–ganglioside complexes present (Extended Data Fig. 5a), were isolated and then collisionally heated to release bound gangliosides (Extended Data Fig. 5b). Notably, signals corresponding to deprotonated GM1, GM2 and GM3 ions were measured; no signals corresponding to the disialylated gangliosides (GD1a, GD2 or GT1b) were detected. As a negative control, these measurements were repeated using identical experimental conditions but in the absence of the RBD, and no ganglioside ions were detected (Extended Data Fig. 5c). Even though GM3 was the most abundant of the gangliosides detected, this may not represent an intrinsic affinity for the RBD but rather a greater release efficiency from the ND as the RBD complex, as suggested from a previous study37.
Inhibition by Sia and HS
The screening and affinity results reveal that the RBD binds preferentially to sialoglycans and HS oligomers as well as other structures, such as ABH antigens (Fig. 1a). The diversity of recognized structures raises the possibility that the RBD (and the S protein) possesses multiple glycan-binding sites with distinct binding properties. Experimentally derived structural data have yet to be reported for RBD–glycan complexes, and the only glycan-binding site (for HS) was predicted from molecular docking4. With the goal of testing whether the RBD possesses multiple glycan-binding sites, we performed CaR-ESI-MS screening of the 16 sublibraries of defined glycans against the RBD in the presence of 20- and 50-fold excess (relative to RBD) of methyl α-sialoside (MeαNeu5Ac) at 25 °C (Supplementary Fig. 5). These conditions are expected to lead to a reduction in the concentration of RBD-binding site(s), at least for those that recognize Sia.
The MeαNeu5Ac concentration-dependent relative abundances of released ligands are summarized in Supplementary Fig. 5d. Inspection of the glycan screening results revealed that at a high MeαNeu5Ac concentration, the most abundant ligand released from RBD was not 71 but rather the neutral pentasaccharide 87 (Galα(1-3)[Fucα(1-2)]Galβ(1-4)[Fucα(1-3)]Glc), which contains a fucosylated B type VI blood group at the terminus. Overall, the relative abundances measured for the neutral glycan ligands increased relative to those of the acidic ligands (Fig. 3a). These data suggest that the RBD possesses at least two glycan-binding sites with distinct preferences for neutral and acidic ligands.
a, Scatter plot of the relative change in normalized abundances (to 71) of released neutral and acidic glycans in the presence and absence of MeαNeu5Ac. Measurements were performed in aqueous ammonium acetate solutions (100 mM, pH 6.9, 25 °C) of RBD (10 μM) and each of the defined libraries (50 nM of each glycan) with 0, 200, and 500 µM MeαNeu5Ac. Circles and diamonds represent neutral and acidic glycans, respectively, and solid lines and dashed lines represent the median values and first and third quartiles, respectively. Statistical significance was calculated based on an unpaired Mann–Whitney test because the distribution of data failed numerous normality tests (D’Agostino–Pearson omnibus, Shapiro–Wilk and Kolmogorov–Smirnov); n = 3 independent experiments for each glycan. b, Change in normalized abundance of released glycan 69 following the addition of glycan 41 or MeαNeu5Ac to the solution. Measurements were performed in aqueous ammonium acetate solutions (100 mM, pH 6.9, 25 °C) of RBD (5 μM) and 69 (50 nM) in the absence and presence of 41 (triangle) or MeαNeu5Ac (circle) at concentrations ranging from 0.1 to 100 μM (41) or 500 μM (MeαNeu5Ac). Ions in the range of m/z 3,000–4,000 were subjected to HCD using a collision energy of 50 V. Data represent mean ± s.d.; n = 3 independent experiments. c, Zero-charge mass spectra of RBD (5 μM) alone and in the presence of glycan 69 (25 μM) or glycan 41 (50 μM) or a mixture of 69 (25 μM) and 41 (50 μM). An asterisk indicates a sodium adduct. Experiments were performed in aqueous ammonium acetate solutions (100 mM, pH 6.9, 25 °C). Deconvolution was performed using the Thermo BioPharma Finder Software.
Next, we considered whether HS and sialoglycans share a common binding site. We performed CaR-ESI-MS on solutions of the RBD and ganglioside 69 at fixed concentrations and varying concentrations of 41 (Fig. 3b). Notably, the relative abundance of released 69 decreased with increasing concentration of 41, consistent with competitive ligand binding. Analogous measurements performed using MeαNeu5Ac (instead of 41) produced qualitatively similar results as 41 (Fig. 3b). We also performed direct ESI-MS measurements on a solution of RBD, 69 and 41 to establish whether the ternary complexes of RBD with 69 and 41 were present in solution. However, analysis of the resulting zero-charge mass spectrum did not reveal evidence of such ternary complexes, further suggesting that HS and sialoglycans share a common binding site (Fig. 3c).
Sia-dependent RBD binding and viral entry
The results above suggest that Sia glycoconjugates are important for infection of cells by SARS-CoV-2. To test this hypothesis, we used flow cytometry-based assays on ACE2-expressing HEK293 cells (Extended Data Fig. 6a). When using a trimeric RBD with a C-terminal mPlum fusion38, we observed robust binding of the SARS-CoV-2 RBD to ACE2+ cells but no binding to ACE2–/– cells (Extended Data Fig. 6b). Pseudotyped SARS-CoV-2 lentivirus encoding green fluorescent protein (GFP) showed robust infection of ACE2+ HEK293 cells 24 h after a 1-h incubation (Extended Data Fig. 6c).
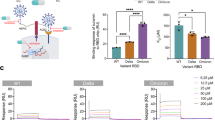
To test the role of Sia in RBD binding and viral entry, we used pharmacological, genetic, and enzymatic approaches to decrease Sia levels. ACE2+ HEK293 cells were first treated with the ST inhibitor 3FNeu5Ac39 or its non-fluorinated analog as a control for 3 d. This substantially decreased Sia on the cell surface, as measured by flow cytometry (Extended Data Fig. 7) with the Sambucus nigra lectin (SNA; binds select sialosides; Fig. 4a) and peanut agglutinin (PNA; binding is inhibited by select sialosides; Fig. 4b). In 3FNeu5Ac-treated cells, RBD binding was consistently decreased (Fig. 4c), and pseudotyped lentivirus showed less infection (Fig. 4d). The decrease in infectivity was approximately 30–40%, which was consistent over numerous independent experiments. Using immunofluorescence staining of Vero-E6 cells, a decrease in RBD binding was observed in cells treated with 3FNeu5Ac (Supplementary Fig. 6). Importantly, ST inhibition did not alter ACE2 levels (Fig. 4e and Supplementary Fig. 11). Using a control lentivirus that does not encode the SARS-CoV-2 S protein, no differences were observed in viral entry following 3FNeu5Ac treatment (Supplementary Fig. 7a).
a,b, Changes in Sia levels from 3FNeu5Ac treatment of HEK293 ACE2+ cells determined by SNA (a) and PNA (b) lectin staining by flow cytometry. These n = 3 biological replicates are represented as mean fluorescence intensity (MFI) values with 95% confidence interval (95% CI) error. c, 3FNeu5Ac treatment decreases RBD binding. The MFI values for n = 3 biological replicates within a single experiment (left) and the average of n = 3 independent experiments (right) are shown. The average RBD binding to control cells was set to 100%. d, 3FNeu5Ac treatment decreases SARS-CoV-2 pseudovirus infection of HEK293 ACE2+ cells. The percentage of GFP+ cells for n = 3 biological replicates within a single experiment (left) and the averages of n = 3 independent experiments (right) are shown. The error bars associated with the single experiment are 95% CI. e, ACE2 expression level does not change following 3FNeu5Ac treatment. The MFI with 95% CI for n = 3 biological replicates is shown. f–j, Flow cytometry results for changes in Sia levels within CMAS–/– ACE2+ HEK293 cells determined by SNA (f) and PNA (g) lectin staining, RBD binding (h), SARS-CoV-2 pseudovirus infection (i) and ACE2 expression (j) on CMAS+ versus CMAS–/– ACE2+ HEK293 cells. Each point represents the average of n = 3 biological replicates for each individual clone with 95% CI error bars. k, SARS-CoV-2 pseudovirus infection of CMAS+ versus CMAS–/– ACE2+ HEK293 cells treated with or without 3FNeu5Ac. The percentage of GFP+ cells for n = 3 biological replicates with 95% CI error bars is shown. l,m, Pretreatment of Vero-E6 cells (l) and ferret lung tissue sections (m) with neuraminidase decreases RBD binding. Scale bars represent 100 μm (l) and 200 μm (m), and results represent data from one representative sample from n = 5 experiments. n, Quantification of fluorescence intensity for RBD binding and lectin staining of Vero-E6 cells and ferret lung tissue sections. Error bars represent ±s.d. of n = 3 replicates. Statistical significance was calculated based on either two-tailed unpaired Student’s t-test (a–k) or analysis of variance (ANOVA) with multiple comparisons (l,m). Flow cytometry data were analyzed on FlowJo version 9.9.6, and graphs were plotted on GraphPad Prism 8; NS, not significant.
We also genetically ablated Sia through isolation of a number of CMP Sia synthetase (CMAS)+ and CMAS–/– clones by CRISPR/Cas9 in ACE2+ HEK293 cells, which were all tested in parallel to minimize concerns of clonal variability. Compared to the 12 isolated CMAS+ clones, the 3 CMAS–/– clones exhibited decreased SNA staining (Fig. 4f) and enhanced PNA staining (Fig. 4g), consistent with the abrogation of CMP Sia biosynthesis. RBD binding was decreased (Fig. 4h), and pseudotyped viral entry was likewise suppressed in the CMAS–/– clones (Fig. 4i), for which there were no significant differences in ACE2 expression levels (Fig. 4j). A single CMAS+ clone and a single CMAS–/– clone with comparable ACE2 expression levels were selected and fed with 3FNeu5Ac. Results showed that treatment with 3FNeu5Ac decreased pseudotyped viral infectivity in the CMAS+ clones, but did not alter viral infectivity in the CMAS–/– clones (Fig. 4k). The lack of an effect of 3FNeu5Ac in CMAS–/– cells strongly suggests that the effects of 3FNeu5Ac on infectivity are due to the lower cell surface Sia levels.
As a third method for reducing Sia levels on cells, Vero-E6 cells (Fig. 4l) and ferret lung tissue sections (Fig. 4m) were fixed and pretreated with neuraminidase from Vibrio cholerae before assessing RBD binding by immunofluorescence staining. SARS-CoV-2 RBD binding was decreased by nearly 80% for both cells and tissue, despite no differences in ACE2 levels, with the expected abrogation of Sia reported by SNA and Maackia amurensis lectin (MAL) staining (Fig. 4n). These results were consistent with three different sources of neuraminidase and required overnight treatment with the neuraminidases (Supplementary Fig. 8). Moreover, abrogating cellular HS by treating cells with heparanase also decreased RBD binding to cells (Supplementary Fig. 9).
Glycolipids are critical for SARS-CoV-2 pseudoviral infection
We next investigated the potential for glycolipids to play a role in RBD binding and viral entry, given that gangliosides were the top hit in our binding assays. We used an inhibitor of UDP-glucose ceramide glycosyltransferase (UGCG), called GENZ-123346. ACE2+ HEK293 cells treated for 2 d with this inhibitor exhibited decreased cholera toxin (CTX) staining by flow cytometry (Extended Data Fig. 8), which is indicative of decreased glycolipid levels on cells (Fig. 5a,b). SARS-CoV-2 RBD binding (Fig. 5c) and pseudotyped viral infection (Fig. 5d) were tested in cells depleted of glycolipids with GENZ-123346, both of which showed significant decreases for the cells treated with the inhibitor. Moreover, GENZ-123346 failed to elicit an effect on pseudoviral entry with CMAS–/– cells (Fig. 5e), suggesting the importance of Sia-containing glycolipids. Infection of a control lentivirus not encoding the SARS-CoV-2 S protein did not show differences in infection following treatment with GENZ-123346 (Supplementary Fig. 7b). Endoglycoceramidase was used as an additional means of destroying cellular glycolipids40, which resulted in the loss of SARS-CoV-2 RBD binding to Vero-E6 cells (Supplementary Fig. 10). Moreover, V. cholerae neuraminidase could not completely abrogate CTX staining of cells after a 1-h treatment, but a 16-h treatment completely abrogated the signal, which was consistent with results presented above in which a 1-h treatment with neuraminidase did not completely abrogate RBD binding (Supplementary Fig. 11).
a,b, Changes in CTX binding on ACE2+ HEK293 cells after GENZ-123346 treatment determined by flow cytometry. Representative histogram results (a) and quantification of the MFI (b) are shown. Error bars represent 95% CI of n = 3 biological replicates. c,d, RBD binding to ACE2+ HEK293 cells (c) and SARS-CoV-2 pseudovirus infection in ACE2+ HEK293 cells (d) following GENZ-123346 treatment; n = 3 biological replicates are shown with 95% CI error bars. e, SARS-CoV-2 pseudovirus infection in CMAS+ and CMAS–/– ACE2+ HEK293 cells following GENZ-123346 treatment. The percentage of GFP+ cells for n = 3 biological replicates and 95% CI error bars are shown. f–j, CTX staining (f), ACE2 expression levels (g), RBD binding (h) and percentage of GFP+ cells (i) of UGCG+ and UGCG–/– ACE2+ HEK293 cell clones. Error bars represent 95% CI of n = 3 biological replicates. j, Pseudoviral infection of ACE2+ UGCG–/– HEK293 cells with supplementation of glycolipids isolated from wild-type HEK293 cells. The percentage of GFP+ cells for n = 4 biological replicates within a single experiment (left) and the averages of n = 3 independent experiments (right) with error bars representing 95% CI are shown. Statistical significance was calculated based on two-tailed unpaired Student’s t-test (b–j). Flow cytometry data were analyzed on FlowJo version 9.9.6, and graphs were plotted on GraphPad Prism 8; NS, not significant.
As a complementary approach, glycolipids were also depleted in cells by genetic ablation of UGCG by CRISPR/Cas9. Three UGCG+ and four UGCG–/– clones were isolated, which stained positive and negative for CTX, respectively (Fig. 5f). Loss of UGCG did not significantly alter expression of ACE2 (Fig. 5g). A single UGCG+ clone and a single UGCG–/– clone were selected with similar ACE2 expression levels for further testing. The UGCG–/– cells showed decreased RBD binding (Fig. 5h) and were infected to a lesser extent by SARS-CoV-2 pseudotyped lentivirus (Fig. 5i). Moreover, glycolipids extracted from UGCG+ HEK293 cells and supplemented into UGCG–/– ACE2+ HEK293 cells rescued pseudoviral infection (Fig. 5j). Overall, these results with genetic and pharmacological ablation of glycolipid biosynthesis suggest an important role for Sia-containing glycolipids in SARS-CoV-2 infection of cells.
Glycolipids are critical for infection by SARS-CoV-2 virus
We next assessed whether infection of two different strains of authentic SARS-CoV-2 virus (hCoV-19/Canada/ON-VIDO-01/2020 (VIDO) and 72B/CA/CALG (72b Calg)) were impacted by glycolipids. A threefold decrease in infection of both strains in ACE2+ HEK293 cells was observed when glycolipids were depleted by GENZ-123346 (Fig. 6a). UGCG–/– HEK293 ACE2-expressing cells showed decreased 72b Calg SARS-CoV-2 infection compared to that of UGCG+ cells and were not affected by GENZ-123346 treatment unlike UGCG+ cells (Fig. 6b). Moreover, infection of primary human nasal epithelial cells decreased following GENZ-123346 treatment (Fig. 6c). These results demonstrate that pharmacological depletion of glycolipids decreases infection of authentic SARS-CoV-2 virus in both cell lines and primary cells.
a,b, SARS-CoV-2 infection by two authentic SARS-CoV-2 viral strains, VIDO and 72b Calg, in ACE2+ HEK293 cells after GENZ-123346 treatment (a) or in UGCG+ or UGCG–/– ACE2+ HEK293 cells (b). Shown is the viral titer of n = 3 biological replicates with 95% CI error bars. c, Infection of GENZ-123346-treated human primary nasal epithelial cells by 72b Calg virus. Open and solid symbols represent n = 2 biological replicates each containing n = 2 technical replicates at 0.1 and 0.5 multiplicity of infection (MOI). Viral titer was quantified by a plaque-forming unit (PFU) assay. Error bars represent 95% CI. Statistical significance was calculated using two-tailed unpaired Student’s t-test (a–c). Graphs were plotted using GraphPad Prism 8; NS, not significant.
Discussion
In this study, we leveraged CaR-ESI-MS, a sensitive and label- and immobilization-free assay, to study defined and natural glycan libraries to identify human glycan structures recognized by SARS-CoV-2 that may facilitate viral infection. Screening a defined glycan library against the SARS-CoV-2 RBD and S protein revealed that several different classes of structures are recognized. Notably, both the RBD and S protein bind a variety of acidic glycans, with the highest preference for the oligosaccharides on the gangliosides GM1 and GM2. Screening of NDs containing a mixture of gangliosides confirmed RBD binding to GM1 and GM2 as well as GM3. We report RBD binding to monosialylated gangliosides embedded in a lipid bilayer. The affinities of the monosialylated ganglioside oligosaccharides for SARS-CoV-2 RBD (for example, 160 ± 40 μM for GM1-pentasaccharide at 25 °C) are similar and, in some instances, stronger than affinities reported for other ganglioside-binding viruses (for example, respiratory syncytial virus and influenza virus strains) for which cell entry is Sia dependent41,42. These millimolar affinities for a monovalent protein–glycan interaction can be enhanced by a virus particle on the cell surface through multivalency/avidity stemming from multiple low-affinity binding sites, resulting in biologically relevant interactions43. It is also noteworthy that, while other members of the coronavirus family also bind glycolipids, they do so through their N-terminal domain; therefore, our observation of binding glycolipids through the RBD is an entirely new finding. Interestingly, no binding of S1 to the glycans of glycosphingolipids, including gangliosides, was detected in glycan microarray screening6. This absence of binding (with the array) might be due to the relatively low affinities of these interactions or deleterious effects of glycan labeling on binding.
SARS-CoV-2 primarily affects the respiratory system, but it can also invade multiple organ systems. Recently, it was revealed that SARS-CoV-2 can invade the central nervous system (CNS)44. Infection of neurons by SARS-CoV-2 is highly relevant to our findings as gangliosides are prominent in the human brain at 10- to 30-fold higher levels than in other organs45. Therefore, gangliosides possibly play an important role in mediating SARS-CoV-2 infection of the CNS.
Acidic HMOs were also observed at high relative abundances in CaR-ESI-MS screening, raising the possibility (based on an assumption that HMOs can act as viral inhibitors) that breastfed newborns may be protected against SARS-CoV-2 infection. Indeed, in a study of 72 neonates breastfed by COVID-19-positive mothers, it was found that none tested positive for infection after 14 d46. Interestingly, some neutral glycans, including ABH blood group antigens (in particular A and H types 2, 3 and 4), were also found to exhibit moderate CaR-ESI-MS signal. Very recently, binding of the RBD to blood group A was demonstrated19, which is consistent with our findings. Together, these findings may offer a clue as to why individuals with blood group A are at higher risk of hospitalization following SARS-CoV-2 infection47. Moreover, based on the competitive binding experiments with MeαNeu5Ac, which competed away many of the sialoglycans but did not perturb binding of the neutral glycans, we suggest that the RBD may have at least two distinct glycan-binding sites.
The affinities measured for HS dp2–dp6 oligosaccharides (39–41), which range from 200 μM to 400 μM (at 25 °C), are similar to the Kd for the RBD and a long-chain heparin sample (240 μM, with MWs ranging from 17 kDa to 19 kDa)6 but weaker than the value for heparin (1 μM; average MW of 15,700 Da)5. Notably, in the former surface plasmon resonance study, it was the RBD that was immobilized, while the glycan was immobilized in the latter. Therefore, simple avidity considerations likely explain this discrepancy in Kd values. Esko and co-workers reported that destruction of cellular glycosaminoglycans (GAGs) decreased SARS-CoV-2 infection, leading to the proposal that GAGs serve as a co-receptor. Notably, the affinities we report for ganglioside binding are as strong, or stronger, than for HS. The addition of HS (41) was shown to reduce the relative abundance of GM1-pentasaccharide (69) released from RBD in CaR-ESI-MS measurements. Moreover, ESI-MS measurements failed to detect evidence of the ternary complexes of the RBD, 69 and 41. Together, these results suggest that sialoglycans and HS bind competitively and presumably share a common binding site. Accordingly, our demonstration that reduction of Sia or ganglioside levels on cells decreases RBD binding and infection of both pseudotyped and authentic SARS-CoV-2 virus argues that sialoglycans may be equally as important as GAGs. Very recently, it was reported by others that, similar to what we observed, inhibition of UGCG diminishes SARS-CoV-2 pseudotyped viral entry48. However, our findings go beyond that by showing direct binding interactions between the RBD and gangliosides, that inhibition of UGCG has no effect in cells lacking Sia and that genetic ablation of UGCG also causes a decrease in viral infection by both pseudotyped and authentic virus. It is worth noting that HEK293 cells, which we used extensively in our experiments, contain predominantly GM3, GM2, GD3 and GT1b along with trace amounts of GM149.
In summary, our findings suggest that sialylated glycans, which are abundant on all human cells, are bound through the RBD of SARS-CoV-2, thereby facilitating viral entry. These findings may have important implications in the tissue tropism of SARS-CoV-2 and could lead to new therapeutic approaches.
Methods
Ethics
The study protocol for acquisition of human tissue samples conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was reviewed by the ethics review board at the University of Alberta (Pro00005105, Pro00085859 and Pro00035875).
The ferret lung tissues used in this study came from ferrets handled in an ABSL3 biocontainment laboratory. The research on these animals was conducted in compliance with the Dutch legislation for the protection of animals used for scientific purposes (2014, implementing EU Directive 2010/63) and other relevant regulations. The licensed establishment where this research was conducted (Erasmus MC) has an approved OLAW Assurance A5051-01. Research was conducted under a project license from the Dutch competent authority, and the study protocol (17–4312) was approved by the Institutional Animal Welfare Body. Animals were housed in class III isolators in groups of two.
Proteins
SARS-CoV-2 S protein50 and RBD (consisting of residues 319–541 and a C-terminal hexa histidine tag)51 were expressed in HEK293 cells and purified as described elsewhere. The lectins from Sambucus nigra (elder; SNA) and Maackia amurensis (MAL), lysozyme from chicken egg white and α-lactalbumin were purchased from Sigma-Aldrich. The P dimer of the Saga strain (GII.4, genotype; MW 69,734 Da) was produced as described elsewhere52. Each protein was dialyzed and concentrated against 100 mM ammonium acetate (pH 6.9) using an Amicon 0.5-ml microconcentrator (EMD Millipore) with a 10-kDa or 30-kDa MW cutoff and stored at –20 °C until use. The membrane scaffold protein (MSP) MSP1E1 (MW 27,494 Da) was expressed from the plasmid pMSP1E1 (Addgene) and purified using a reported protocol53. The MSP1E1 protein was dialyzed into Tris-HCl buffer (10 mM Tris, 100 mM NaCl, 1 mM EDTA, pH 7.4) and stored at –80 °C until needed. The concentrations of each stock solution were estimated by UV absorption at 280 nm.
Defined glycan library
The structures, MWs and sources of the purified oligosaccharides contained within the defined library are shown in Extended Data Fig. 1 and Supplementary Fig. 1 and are summarized in Supplementary Table 1. Glycans 1, 2–5, 7–14, 33–38, 42–76, 78–82, 84, 87–90, 92–102, 104, 108, 111, 112, 115, 121, 126 and 128 were purchased from Elicityl SA, glycans 6, 77, 83, 85, 86, 91, 116, 117, 124 and 130 were purchased from Dextra, glycans 103 and 122 were purchased from Carbosynth and glycans 105–107, 109, 110, 113, 114, 118–120, 123, 125, 127, 129, 131 and 132 were purchased from IsoSep. Glycans 15–32, 39–41 and 133–139 were prepared as described elsewhere. The N-glycan 140 was purchased from Chemily Glycoscience. Truncated ceramide functionalized GM1-pentasaccharide (MW 1,123.40 Da) was a gift from D.R. Bundle (University of Alberta).
With the exception of 39–41 and 140, the glycans were divided into 16 MW-unique sublibraries (libraries A–P). To prepare the sublibraries, 1 mM stock solutions of each glycan were first prepared by dissolving a known mass of glycan in ultrafiltered Milli-Q water. Aliquots of these solutions were then mixed with ultrafiltered Milli-Q water to give 50 μM stock solutions of each sublibrary. All stock solutions were stored at –20 °C until needed.
Phospholipid and glycosphingolipids
The gangliosides GM1 (β-d-Gal-(1→3)-β-d-GalNAc-(1→4)-[α-d-Neu5Ac-(2→3)]-β-d-Gal-(1→4)-β-d-Glc-ceramide, major isoforms d18:1-18:0 with MW 1,545.88 Da and d20:1-18:0 with MW 1,573.91 Da), GM2 (β-d-GalNAc-(1→4)-[α-d-Neu5Ac-(2→3)]-β-d-Gal-(1→4)-β-d-Glc-ceramide, major isoforms d18:1-18:0 with MW 1,383.82 Da and d20:1-18:0 with MW 1,411.86 Da) and GM3 (α-d-Neu5Ac-(2→3)-β-d-Gal-(1→4)-β-d-Glc-ceramide, major isoforms d18:1-18:0 with MW 1,180.74 Da and d20:1-18:0 with MW 1,208.78 Da) were purchased from Cedarlane Labs. GD1a (α-d-Neu5Ac-(2→3)-β-d-Gal-(1→3)-β-d-GalNAc-(1→4)-[α-d-Neu5Ac-(2→3)]-β-d-Gal-(1→4)-β-d-Glc-ceramide, major isoforms d18:1-18:0 with MW 1,836.97 Da and d20:1-18:0 with MW 1,865.00 Da) and GT1b (α-Neu5Ac-(2→3)-β-d-Gal-(1→3)-β-d-GalNAc-(1→4)-[α-Neu5Ac-(2→8)-α-Neu5Ac-(2→3)]-β-d-Gal-(1→4)-β-d-Glc-ceramide, major isoforms d18:1-18:0 with MW 2,128.07 Da and d20:1-18:0 with MW 2,156.10 Da) were purchased from Sigma-Aldrich. GD2 (β-d-GalNAc-(1→4)-[α-d-Neu5Ac-(2→8)-α-d-Neu5Ac-(2→3)]-β-d-Gal-(1→4)-β-d-Glc-ceramide, major isoforms d18:1-18:0 with MW 1,674.92 Da and d20:1-18:0 with MW 1,702.95 Da) was purchased from MyBioSource. The phospholipid DMPC (MW 677.50 Da) was purchased from Avanti Polar Lipids (Supplementary Fig. 12). Stock solutions of each ganglioside (1 mM) and DMPC (20 mM) were prepared in HPLC-grade methanol/chloroform (1:1 (vol/vol); Thermo Fisher).
Synthesis of methyl (5-acetamido-4,7,8,9-tetra-O-acetyl-3-dehydro-3,5-dideoxy-3-fluoro-5-α-d-glycero-d-galacto)onate (142) and methyl (5-acetamido-4,7,8,9-tetra-O-acetyl-2-methoxy-2,3,5-tri-deoxy-d-glycereo-β-d-galacto-2-nonulopyranosid)onate (145)
Additional details for preparation of these compounds are described in the Supplementary Notes.
Preparation of the protein pellet from lung and intestinal tissues
Two natural libraries of N-glycans prepared from intestinal tissue (from an individual with Crohn’s disease) and lung tissue were screened against RBD. Lung tissue was taken after ex vivo perfusion of human lung (‘right, peripheral’) from a non-COVID-19-related human donor and was snap-frozen and stored in liquid nitrogen.
Protein extraction from lung or intestinal tissue was performed as described below. Briefly, the collected sample was snap-frozen immediately and stored at –80 °C until homogenization in ice-cold water using a PowerGen 125 homogenizer (Fisher Scientific). Then, 2.7 times the aqueous sample volume of methanol and 1.3 times the aqueous sample volume of chloroform were sequentially added to the homogenized sample and mixed vigorously at room temperature. The homogenate was centrifuged at 3,000g for 10 min followed by removal of the supernatant. The excess methanol/chloroform from the pellet was removed under a stream of nitrogen gas for 2 min. After addition of 0.6 M Tris buffer (pH 8.5, adjusted with dilute acetic acid), the sample was placed under a stream of nitrogen and evaporated. The sample was then resuspended in aqueous 200 mM ammonium acetate solution followed by centrifugation at 4,000 r.p.m. for 10 min. The supernatant was lyophilized and stored at –20 °C until needed.
N-glycan sample preparation
To prepare the N-glycan libraries, 400 μg of SARS-CoV-2 RBD or 1 g of protein pellet extracted from lung or intestinal tissue was dissolved in 500 μl of 8 M urea in 100 mM Tris-HCl (pH 8.0) containing 3 mM EDTA and incubated at room temperature for 1 h. The denatured RBD was then reduced with 10 µl of 500 mM dithiothreitol (DTT; Sigma-Aldrich) at room temperature for 1 h followed by alkylation with 23 µl of 500 mM iodoacetamide (Sigma-Aldrich) at room temperature for 20 min in the dark. The reaction was quenched by adding 10 µl of 250 mM DTT, and the solution buffer was exchanged using a PD MiniTrap G-25 column (GE Healthcare) according to the manufacturer’s instructions. The glycoprotein solution was subsequently digested with trypsin/chymotrypsin (substrate/enzyme (wt/wt) = 50) in 50 mM ammonium bicarbonate (pH 8.0) for 18 h at 37 °C. The reaction was quenched by heat inactivation at 100 °C for 10 min.
Released N-glycans were liberated by incubation of resulting glycopeptides with amidase PNGase F (New England Biolabs) at 37 °C for 18 h and purified using porous graphitized carbon (Hypercarb cartridges, 100 mg, 1-ml volume; Thermo Fisher Scientific). The porous graphitized carbon cartridge was preequilibrated with 1 ml of 80% acetonitrile containing 0.1% trifluoroacetic acid (TFA) followed by 2 ml of water. The samples were added to the cartridges followed by washing with 2 ml of water. N-glycans were then eluted with 1 ml of 25% acetonitrile containing 0.1% TFA. The lyophilized sample was stored at –20 °C until needed.
Labeling of N-glycan libraries
The N-glycans released from RBD, lung or intestinal tissue were fluorescently labeled via reductive amination with 2-AB (Sigma-Aldrich). The 2-AB and sodium cyanoborohydride were dissolved in 70%:30% (vol/vol) DMSO:acetic acid to provide concentrations of 0.37 and 1 M, respectively. The labeling solution was added to the N-glycan sample followed by incubation at 65 °C for 3 h. The excess labeling reagent was removed by a 30-mg sorbent Oasis HLB cartridge (Waters). The cartridge was conditioned by 2 × 1 ml of acetonitrile. The labeled N-glycans were diluted tenfold with acetonitrile and added to a conditioned cartridge. The loaded cartridge was washed two times with 1 ml of acetonitrile and eluted with 2 × 400 µl of 20% acetonitrile in water. The purified N-glycans were lyophilized and stored at –20 °C before analysis.
Preparation of ganglioside-containing NDs
An ND composed of DMPC and a mixture of GM1, GM2, GM3, GD1a, GD2 and GT1b (each nominally 1% of total lipid) was prepared according to the protocol described by Sligar and co-workers53. Briefly, DMPC and the six gangliosides were diluted in methanol at the desired molar ratios. The lipids were dried under a gentle stream of nitrogen to form a lipid film and were then resuspended in a buffer (pH 7.4) of 20 mM Tris-HCl, 0.5 mM EDTA, 100 mM NaCl and 25 mM sodium cholate (Sigma-Aldrich). The MSP MSP1E1 was then added to the mixture at a 1:100 (MSP1E1:(gangliosides + DMPC)) molar ratio. The ND self-assembly process was initiated by adding prewashed Bio-Beads (Bio-Rad), and the mixture was incubated for 3 h at room temperature using an orbital shaker. Following incubation, the supernatant, which contained the NDs, was removed and purified by gel-filtration chromatography using a Superdex 200 10/300 size-exclusion column (GE Healthcare Life Sciences) equilibrated with 200 mM ammonium acetate (pH 6.8). Finally, the fraction corresponding to the NDs was collected, concentrated, dialyzed into 200 mM ammonium acetate (pH 6.8) using an Amicon microconcentrator (EMD Millipore) with a 30-kDa MW cutoff and stored at –20 °C until use. The concentrations of the ND stock solutions were estimated from the concentration of MSP1E1, which was measured by UV absorption at 280 nm and taking into account that each ND contains two copies of MSP1E1.
Mass spectrometry
The CaR-ESI-MS glycan library screening measurements were performed in negative mode using a Q Exactive UHMR Orbitrap mass spectrometer (Thermo Fisher Scientific). The capillary temperature was 200 °C, and the S-lens RF level was 100; an automatic gain control target of 5 × 105 and maximum injection time of 200 ms were used. The resolving power for full MS and HCD were set to 3,125 and 25,000, respectively. HCD spectra were acquired using collision energies ranging from 10 V to 300 V. The ESI-MS affinity measurements were performed in positive ion mode on a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific). The capillary temperature was 200 °C, and the S-lens RF level was 100; an automatic gain control target of 5 × 105 and maximum injection time of 200 ms were used. The resolving power was 17,500. For both instruments, data acquisition and preprocessing was performed using Xcalibur version 4.1; ion intensities were extracted using in-house software (SWARM)54. The CaR-ESI-MS analysis of ganglioside NDs was performed in negative ion mode using a Synapt G2S quadrupole–ion mobility separation–time of flight (Q–IMS–TOF) mass spectrometer (Waters). The source temperature was 60 °C, the cone voltage was 60 V, the trap voltage was 5 V (for MS) or 50 V (for CID) and the transfer voltage was 2 V. Argon was used for collision-induced dissociation (CID) at a Trap ion guide pressure of 1.42 × 10–2 mbar. Data acquisition and processing were performed using MassLynx software (version 4.1).
All three instruments were equipped with a modified nanoflow ESI (nanoESI) source. NanoESI tips with an outer diameter (o.d.) of ∼5 µm were pulled from borosilicate glass (1.0-mm o.d., 0.78-mm inner diameter) with a P-1000 micropipette puller (Sutter Instruments). A platinum wire was inserted into the nanoESI tip, making contact with the sample solution. A voltage of approximately 1 kV (positive mode) or –1 kV (negative mode) was applied to the platinum wire. The temperature of the solution in the nanoESI tip was controlled using a home-built temperature-controlled device55.
CaR-ESI-MS library screening
For all screening experiments, aqueous solutions composed of the volatile ammonium acetate salt (100 mM, pH 6.9), RBD (10 μM) or S protein (280 μg ml–1) and sublibrary (50 nM (RBD) or 500 nM (S protein) of each glycan) were used. The SNA lectin, which has strict preference for α2–α6-linked sialosides, served as Pref for libraries containing neutral glycans and those containing Neu5Acα2-3 and Neu5Acα2-8 (libraries A–M, O and P), and MAL, which is specific for certain α2–α3-linked sialosides, was used for library N, which contained Neu5Acα2-6 glycans. Due to overlap of ion signal corresponding to S protein and SNA and MAL, a Pref was not used; instead, library P served as a negative control for S protein screening. Additionally, the truncated ceramide functionalized GM1-pentasaccharide (Lref) was added to each library as a positive control to rank affinities of released glycan ligands present in each sublibrary. RBD–ligand complex ions with m/z values in the range of 3,000–4,000 were isolated for HCD; S protein–ligand complex ions with m/z values in the range of 8,000–15,000 were isolated. Nitrogen gas and a range of HCD energies (10–60 V) were used to release ligands from the RBD–ligand complexes. The onset of ligand release was observed at energies between 22 V and 32 V. Although there were a few exceptions, the trends in relative abundances of the released ligands were similar at collision energies up to ~50 V (Supplementary Fig. 13). Also, the absolute signal for released ligands increased with energy up to 50 V. At higher energies, the abundances of released ligands were found to decrease due to secondary fragmentation. Consequently, 50 V was selected as the optimal collision energy for library screening. The reproducibility of the CaR-ESI-MS screening results was assessed using library D (Supplementary Fig. 14). The relative standard deviations of the 18 glycan ligands in the library ranged from 2% to 27% (four replicates). Moreover, the trend in relative abundances of released ligands remained constant across the four measurements. Due to the higher MW of the S protein (~550 kDa), argon served as the collision gas, and an optimal collision energy of 300 V was used to release ligands.
CaR-ESI-MS affinity ranking
To rank affinities of released ligands from RBD identified across all sublibraries, the relative abundances (Abrel) of ligands were calculated from equation (1a)
where Lj is a given glycan in the defined library, Ab(Lj) is the total charge-normalized abundance of the deprotonated ions of Lj and any adducts (for example, chloride and nitrate adduct) detected and AbP33 is the total, charge-normalized abundance of the most abundant RBD glycoform (P33; MW 31,926 Da). To rank affinities of glycans released from S protein, the relative abundances (Abrel) of ligands were calculated from equation (1b)
where Ab(Lref) is the charged-normalized abundance of deprotonated Lref ions.
ESI-MS affinity measurements
Kd values of glycan ligands for RBD were measured by the direct ESI-MS binding assay30. As the number of RBD glycan-binding sites is not known, and no RBD species with multiple bound ligands were detected, the ESI-MS data were analyzed assuming a single site. For a monovalent protein–ligand (PL) interaction (equation (2)), the affinity (Kd; equation (3)) can be calculated from the ratio (R) of total Ab of L bound to free P ions (equation (3)) measured by ESI-MS
where [P]0 and [L]0 are initial concentrations of P and L, respectively. The abundance ratio R measured by ESI-MS is taken to be equal to the equilibrium concentration ratio in the solution. Because RBD consists of multiple species with distinct glycan compositions (referred to here as Px, where x = 1–77), and glycosylation can, in principle, influence binding, the affinities for L binding to individual RBD species (equations (5a) and (5x)) were determined.
The corresponding equations of mass balance equations are
where [Px]0 is the initial concentration of a given Px species. Initial concentrations of individual RBD species were estimated from their relative abundances measured by ESI-MS, assuming uniform response factors (equation (7)).
The affinity of a given Px species (Kdx) was calculated from equation (8)
where Rx is the total abundance ratio of ligand-bound and free Px ions (equation (9)).
When detected, the contribution of non-specific Px–L binding (formed during ESI) to the mass spectrum was corrected using the Pref method30.
In some instances, signal for one ligand-bound Px complex overlapped with signal for another (free) Px species. Spectral overlap was corrected for by considering the abundance ratio of the two Px species (for example Px and Px + 1) in the absence of L (r; equation (10)) and assuming that Px and Px + 1 exhibit identical affinities for L.
The corresponding r value was then used to calculate the true Rx value (Rx,cor; equation (11)) corrected for spectral overlap,
which was used in equation (8) to calculate Kdx. Notably, there is good agreement between Rx values measured in the absence of spectra overlap and those corrected for overlap, which indicates that glycosylation has, at best, a minor influence on glycan ligand binding.
Hydrophilic interaction-ultra high performance liquid chromatography analysis of natural N-glycan libraries
The labeled N-glycans released from RBD and the intestinal and lung tissue were analyzed by HILIC-UHPLC on a Thermo Scientific Vanquish UHPLC system coupled with fluorescent (FLD) (Thermo Scientific) and ESI-MS detectors (Thermo Q Exactive Orbitrap). Compound separation was achieved using a Waters Acquity UPLC BEH glycan column (1.7 µm, 2.1 × 150 mm; Waters). The eluents were ammonium formate 100 mM (pH 4.5) (solvent A) and acetonitrile (solvent B). The separation was performed at 60 °C. The following gradient was used for both FLD and MS detection: t = 0 min, 25% solvent A (0.2 ml min–1); t = 175 min, 35% solvent A (0.2 ml min–1); t = 210 min, 45% solvent A (0.2 ml min–1); t = 210.1 min, 100% solvent A (0.2 ml min–1); t = 224 min, 100% solvent A (0.2 ml min–1); t = 225 min, 25% solvent A (0.2 ml min–1); t = 240 min, 25% solvent A (0.2 ml min–1). The excitation and emission wavelengths were set at 330 nm and 420 nm, respectively. During LC–MS analysis, the following parameters were used: probe heater temperature of 250 °C, sheath gas flow rate of 40 arbitrary units (AU), aux gas flow rate of 10 AU, capillary temperature of 275 °C and spray voltage of 3.5 kV. The mass spectra were acquired in positive mode with an m/z range of 250–3,000 at a resolution of 70,000. The automatic gain control target was set at 1 × 106, and a maximum injection time of 100 ms was used. HCD mass spectra were acquired in the data-dependent mode for the five most abundant ions with a resolution of 17,500. Automatic gain control target, maximum injection time and isolation window were set at 2 × 105, 200 ms and 2.0 m/z, respectively. HCD-normalized collision energy was 27%. The data were recorded by Xcalibur (Thermo, version 4.1) and interpreted using GlycoWorkbench software56.
The glycan composition corresponding to each peak identified by HILIC-UHPLC analysis was preliminarily assigned as previously reported23. Briefly, the fractional abundance (Fj) of a given labeled N-glycan within the mixture was determined from the relative fluorescent signal in the chromatogram (equation (12))
where AFj is the peak area of glycan j. In cases where a chromatographic peak contained multiple N-glycan species with distinct MWs, analysis of corresponding extracted ion chromatograms enabled the determination of the fractional abundance (fj) of individual glycans (or mixture of compositional isomers) that comprise a given peak (equation (13))
where Aj is the peak area of the monoisotopic ion of glycan j (summed over all charge states, n), and Aj,coeluting is the sum of all monoisotopic ion peak areas corresponding to coeluting N-glycan species. The fractional abundance of each glycan j (AFj) within the chromatographic peak was calculated using equation (14)
where AFj,coeluting corresponds to the total fluorescent signal of coeluted N-glycans.
Affinity ranking of N-glycan ligands detected by CaR-ESI-MS screening
The ligand affinities (individual glycans or mixtures of compositional isomers) for RBD detected by CaR-ESI-MS were ranked based on their charge-normalized fractional abundances (FAbj) and fractional abundances determined by HILIC-UHPLC (Fj). For each glycan ligand (or mixture), the relative affinity (Krel,j) was calculated using equation (15).
The ligand affinity ranking values (rj) reported for RBD correspond to the Krel,j values normalized to the maximum value Krel,max (equation (16)).
Assignment of glycan compositions of RBD glycoforms
The data were analyzed using the measured MWs of intact protonated RBD. The MW of deglycosylated RBD was calculated based on the elemental composition (C1170H1744N318O335S9) corresponding to residues 319–541 of the S protein (UniProt number P0DTC2) plus the hexa histidine tag and four disulfide bonds. Possible glycan compositions were simulated for the numbers of N-acetylhexosamines (N: HexNAc, N-acetylgalactosamine and N-acetylglucosamine), hexoses (H: Hex, glucose and galactose), fucoses (F: Fuc) and N-acetylneuraminic acids (S: Neu5Ac). Possible values of N, H, F and S were calculated by considering reported O-glycans and the composition of N-glycans established by HILIC-UHPLC-FLD/ESI-MS (Supplementary Tables 3 and 4)32,33,34,35. Possible MWs of RBD glycoforms were then calculated from the sum of aforementioned MW of RBD and MWs of glycan residues from each possible N-H-F-S combination (Source Data Fig. 3).
Flow cytometry
A five-laser LSRFortessa X-20 flow cytometer (BD Biosciences) was used for all flow cytometry data collection. Data were processed using FlowJo software version 9.9.6.
Production of pseudovirus
GFP-encoding SARS-CoV-2 pseudotyped lentivirus was produced from HEK293T cells as reported in ref. 57 using a plasmid encoding GFP (pCCNanoLuc2AeGFP), an envelope plasmid encoding SARS-CoV-2 S protein (SARS-CoV-2) and a packaging plasmid encoding GagPol expression (pCRV1-NL-gagpol). Specifically, 1 × 106 HEK293T cells were plated in a six-well dish with 1.5 ml of DMEM growth medium (Gibco) containing 10% fetal bovine serum (FBS; Gibco), 100 U ml–1 penicillin (Gibco) and 100 µg ml–1 streptomycin (Gibco). The following day, pCCNanoLuc2AeGFP, pCRV1-NL-gagpol and SARS-CoV-2 were mixed in a ratio of 0.7:0.7:0.4 with Opti-MEM medium (Gibco) to give 275 µl. TransIT-LT1 reagent (Mirus Bio) was added according to the manufacturer’s protocol. Cells were incubated with this transfection mixture at 37 °C and 5% CO2 for 72 h. Following transfection, the supernatant was collected, spun at 1,500g at 4 °C for 5 min and incubated at 4 °C in Lenti-X Concentrator (Clontech) for 1 h. The supernatant solution was spun at 1,500g for 45 min at 4 °C. The final pellet was resuspended in one-tenth of the original volume of medium using sterile PBS, titered and stored at –80 °C.
Pseudovirus transduction
HEK293T cells (150,000) expressing ACE258 were plated in triplicate in a 24-well plate in 250 µl of DMEM growth medium (Gibco) containing 10% FBS (Gibco), 100 U ml–1 penicillin (Gibco), 100 µg ml–1 streptomycin (Gibco) and 5 µg ml–1 blasticidin (InvivoGen). Concentrated pseudovirus (10×; 10 µl) was added to each of the 24 wells and incubated for 1 or 8 h at 37 °C and 5% CO2. Cells were washed one time in DMEM medium and were replated in the 24-well plate for 23 or 16 h to allow for expression of the GFP reporter gene. Cells were removed from the plate via trypsin digest, centrifuged (300g, 5 min) and resuspended in flow cytometry buffer (HBSS, 1% FBS and 500 µM EDTA). The viral titer of pseudovirus was determined by examining the percentage of GFP+ cells by flow cytometry.
Production of control lentivirus
An empty lentiviral vector encoding mAmetrine was made using the lentiviral backbone RP172.
Control lentivirus transduction
HEK293T cells (150,000) expressing ACE258 were plated in triplicate in a 24-well plate in 250 µl of DMEM growth medium (Gibco) containing 10% FBS (Gibco), 100 U ml–1 penicillin (Gibco), 100 µg ml–1 streptomycin (Gibco) and 5 µg ml–1 blasticidin (InvivoGen). Concentrated control lentivirus (10×; 10 µl) was added to each of the 24 wells and incubated for 1 h at 37 °C and 5% CO2. Cells were washed one time in DMEM medium and were replated in the 24-well plate for 23 or 16 h to allow for expression of the GFP reporter gene. Cells were removed from the plate via trypsin digestion, centrifuged (300g, 5 min) and resuspended in flow cytometry buffer (HBSS, 1% FBS and 500 µM EDTA). The viral titer of lentivirus was determined by examining the percentage of mAmetrine+ cells by flow cytometry.
Lectin, CTX and anti-ACE2 antibody staining by flow cytometry
Approximately 50,000 cells were seeded in a 96-well U-bottom flask and centrifuged at 300g for 5 min. The cell pellets were resuspended in a solution of HBSS containing calcium and magnesium (Thermo Fisher) and 0.1% bovine serum albumin (BSA) (flow buffer) with either fluorescein-conjugated SNA (1:1,000; Vector Laboratories) or PNA (1:1,000; Vector Laboratories) and incubated on ice for 20 min. After incubation, the cells were washed, centrifuged (300g for 5 min), resuspended in flow buffer and analyzed by flow cytometry. The cells were stained with fluorescein isothiocyanate (FITC)-conjugated CTX (1:1,000; Sigma-Aldrich) at 1 μg ml–1 in flow buffer for 30 min at 4 °C. The cells were washed, resuspended in flow buffer and analyzed by flow cytometry. For ACE2 staining, cells were resuspended in goat anti-human ACE2 antibody (1:500; R&D Systems) in flow buffer. After a 20-min incubation at 4 °C, cells were centrifuged at 300g for 5 min. The pellets were resuspended in a solution of anti-goat AlexaFluor647 secondary antibody (Thermo Fischer) and incubated on ice for another 30 min. Cells were centrifuged at 300g for 5 min, and the cell pellets were resuspended in flow buffer. Differences in the MFI of AlexaFluor647 were measured on the flow cytometer.
Fluorescent SARS-CoV-2 RBD trimer binding by flow cytometry
SARS-CoV-2 293T trimer containing the mPlum fluorophore was tested for cell binding at a final concentration of 10 µg ml–1 in DMEM containing 10% FBS. Cells were incubated at 4 °C for 45 min and centrifuged at 300g for 5 min. The cell pellets were resuspended in flow buffer and analyzed by flow cytometry for mPlum fluorescence in the PE-Cy5 channel.
Extraction of glycolipids from cells
Approximately 6 × 107 HEK293 ACE2 UGCG+ cells were removed from the surface of the flask by trypsin for 2 min. Trypsin was inactivated through the addition of medium (DMEM, 10% FBS), and cells were centrifuged at 300g for 5 min. Cell pellets were resuspended in 5 ml of PBS and centrifuged at 300g for 5 min. Cellular lipids were extracted from the pellet following a procedure described previously59. Briefly, the pellet was resuspended in 1 ml of water. Then, 3.75 ml of methanol:chloroform (2:1) was added to the cell suspension, and the mixture was stirred at room temperature overnight. The solution was centrifuged at 300g for 10 min, the supernatant was set aside, and the pellet was resuspended in 3 ml of methanol:chloroform:water (2:1:0.8). The solution was stirred at room temperature for 2 h and centrifuged at 300g for 10 min. The supernatant was combined with previous supernatants and dried under nitrogen gas, and 1 ml of methanolic sodium hydroxide was added and stirred at 37 °C for 1 h. Solvent was removed under nitrogen gas, and the remaining lipids were resuspended in 1 ml of water and desalted on a G-25 column using water as the eluent. The void volume was collected and freeze-dried in a tared tube.
Supplementation of glycolipids into UGCG–/– HEK293 cells
Approximately 75,000 UGCG–/– HEK293 ACE2+ cells were plated in a 96-well flat-bottom plate. Glycolipid (500 µM) in serum-free DMEM medium was added to wells and incubated at 37 °C for 1 h. After incubation, the medium was removed, cells were resuspended in medium (DMEM, 10% FBS), and the virus was added to each well for 1 h (37 °C, 5% CO2). Cells were washed with DMEM medium and replated in the 96-well plate for a remaining 23 h to allow for expression of the GFP reporter gene. Cells were removed from the plate via trypsin digest, centrifuged at 300g for 5 min and resuspended in flow cytometry buffer (HBSS, 1% FBS and 500 µM EDTA). The viral titer of pseudovirus was determined by examining the percentage of GFP+ cells using flow cytometry.
Immunofluorescent cell staining
Vero-E6 cells grown on coverslips were analyzed by immunofluorescent staining. Cells were fixed with 4% paraformaldehyde in PBS for 25 min at room temperature, after which permeabilization was performed using 0.1% Triton X-100 in PBS. Subsequently, the coronavirus S proteins were applied at 50 μg ml–1 for 1 h at room temperature. Primary strep-monoclonal antibody classic chromeo-555 (IBA) and secondary AlexaFluor488 or AlexaFluor555 goat anti-mouse antibodies (Invitrogen) were applied sequentially with PBS washes in between. DAPI (Invitrogen) was used as a nuclear stain. Samples were imaged on a Leica DMi8 confocal microscope equipped with a ×10 HC PL Apo CS2 objective (0.40 NA). Excitation was achieved with a diode 405 or white light for excitation of Alexa488 and Alexa555, a pulsed white laser (80 MHz) was used at 488 nm and 549 nm, and emissions were obtained in the range of 498–531 nm and 594–627 nm, respectively. Laser powers were 10–20% with a maximum gain of 200. LAS Application Suite X was used as well as ImageJ for the addition of the scale bars. Where indicated, cells were treated with 2 mU of V. cholerae (Sigma, 11080725001) or Arthrobacter ureafaciens (Sigma, 10269611001 or NEB, P0722) neuraminidase in 10 mM potassium acetate and 2.5 mg ml–1 Triton X-100, pH 4.2, at 37 °C overnight.
Tissue staining
Serial sections of formalin-fixed, paraffin-embedded ferret lungs were obtained from the Department of Veterinary Pathobiology at Utrecht University and the Department of Viroscience at Erasmus University, respectively. Tissue was deparaffinized in xylene and rehydrated in a series of alcohol from 100%, 96% to 70% and lastly in distilled water. Tissue slices were boiled in citrate buffer, pH 6.0, for 10 min at 900 kW in a microwave for antigen retrieval and washed in PBS and 0.1% Tween 20 (PBS-T) three times. The tissue slices were incubated with 3% BSA in PBS-T overnight at 4 °C. The next day, purified viral S proteins (50 μg ml–1) or antibodies were added to the tissues for 1 h at room temperature. With rigorous washing steps in between, the secondary antibodies were applied for 45 min at room temperature. Where indicated, tissue slides were treated with 2 mU of V. cholerae neuraminidase in 10 mM potassium acetate and 2.5 mg ml–1 Triton X-100, pH 4.2, at 37 °C overnight.
Pharmacological depletion of Sia
Stocks of peracetylated Neu5Ac or 3FNeu5Ac were prepared in cell culture-grade DMSO at 300 mM and stored at –20 °C. Cells (100,000) in 250 µl of medium were seeded in a 24-well plate. The compound stocks were added to wells at a 1:1,000 dilution to achieve a final concentration of 300 μM. Cells were incubated at 37 °C at 5% CO2 for 72 h to deplete cell surface Sia levels.
Generation of CMAS–/– and UGCG–/– cells
CRISPR RNA (crRNA) was designed to target human CMAS (TGAGACGCCATCAGTTTCGA, Integrated DNA Technologies (IDT)) or human UGCG (CCGATTACACCTCAACAAGA, IDT). HEK293T cells (500,000) expressing ACE2 were plated in a six-well tissue culture plate in 1.5 ml of medium. Twenty-four hours later, guide RNA (gRNA) (1 µM crRNA, 1 µM ATTO-550-labeled transactivating crRNA (tracrRNA; IDT)) was boiled at 95 °C for 5 min. A solution of 20 pmol gRNA, 20 pmol Cas9 nuclease (IDT), 8 µl of Cas9 PLUS reagent (IDT) and 16 µl of CRISPRMAX reagent (Thermo Fisher) in 600 µl of Opti-MEM medium (Gibco) was added to cells and incubated at 37 °C and 5% CO2 for 24 h. The cells were removed from the six-well plate via trypsin digestion, centrifuged (300g, 5 min) and resuspended in 400 µl of FACS buffer (HBSS containing 1% FBS and 500 µM EDTA). The top 5% ATTO-550+ cells were sorted on a BD FACSMelody cell sorter into 96-well flat-bottom plates containing 200 μl of medium at one cell per well and stored at 37 °C and 5% CO2. Approximately 2 weeks later, colonies were screened for CMAS–/– cells on a flow cytometer using fluorescein-conjugated SNA and PNA, while UGCG–/– cells were screened by FITC-conjugated CTX (1:1,000; Sigma-Aldrich). UGCG–/– cells were verified to contain an insertion–deletion (indel) at the correct location by Sanger sequencing.
Glycolipid depletion
Stocks of glucosylceramide synthase inhibitor (GENZ-123346, Toronto Research Chemicals) in cell culture-grade DMSO were prepared at 5 mM and stored at –20 °C. Cells (100,000) in 250 µl of medium were seeded in a 24-well plate. The GENZ-123346 stocks or DMSO as a negative control were added to wells at a 1:1,000 dilution to achieve a final concentration of 5 μM. Cells were incubated with GENZ-123346 at 37 °C and 5% CO2 for 48 h to deplete glycolipids.
Heparanase treatment of ACE2 cells
Heparanase treatment was performed as previously described5. In brief, cells were incubated for 16 h at 37 °C with 0.2 μg ml–1 heparanase. Heparanase was expressed in HEK293T cells using Addgene plasmid 53407.
SARS-CoV-2 viral infection assays
All experiments with SARS-CoV-2 virus were performed under biosafety level 3 (BSL3) conditions. ACE2+ HEK293 cells were cultured as described above. Primary human nasal epithelial cells were obtained from a commercial supplier (Promo Cell) and cultured according to the manufacturer’s recommendations. SARS-CoV-2 (hCoV-19/Canada/ON-VIDO-01/2020, GISAID accession EPI_ISL_425177) was kindly provided by D. Falzarano (Vaccine and Infectious Disease Organization). A D614G-containing strain of SARS-COV-2 (72B/CA/CALG) used in this study was isolated from an individual in Calgary, Alberta, Canda.
Cells (ACE2+ HEK293 or human nasal epithelial cells) were seeded in 96-well plates (1 × 104 cells per well), and the next day, cells were treated with 5 µM GENZ-123346 or DMSO (0.1% final concentration) as a control. After 48 h, cells were infected with the indicated strains of SARS-CoV2 using the indicated MOIs. The virus-containing inoculum was removed after 1 h and replaced with complete fresh medium. Cells were incubated for an additional 24 h before collection of the medium for a plaque assay. Briefly, approximately 1 × 105 cells were seeded in a 24-well plate. After a 24-h incubation, cells were infected with serial dilutions of virus-containing medium (0.2 ml) for 1 h with occasional rocking, after which 1.0 ml of medium containing 0.75% carboxymethyl cellulose was added to the wells. Cells were incubated for 72–96 h to allow plaque formation, after which samples were fixed with 10% formaldehyde, washed with water and stained with 1% crystal violet in 20% ethanol. After extensive washing with water, plaques were counted, and viral titers (PFU per ml) were determined.
Statistics and reproducibility
Unless otherwise indicated, a two-tailed unpaired Student’s t-test was used to assess statistical significance. For experiments with more than two groups, a one-way ANOVA was performed to determine if there were differences between the means. If applicable, the Tukey multiple comparisons post hoc test was used to indicate significance between groups. A P value of <0.05 was accepted as statistically significant. All assays were conducted with replicates of a minimum of n = 3. Values are expressed as the mean ± s.d.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information files. Please contact the corresponding authors for questions regarding the raw data. Additional information will be made available upon reasonable request. Source data are provided with this paper.
References
Maginnis, M. S. Virus-receptor interactions: the key to cellular invasion. J. Mol. Biol. 430, 2590–2611 (2018).
Varki, N. M. & Varki, A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab. Invest. 87, 851–857 (2007).
Matrosovich, M., Herrler, G. & Klenk, H. D. Sialic acid receptors of viruses. Top. Curr. Chem. 367, 1–28 (2015).
Kim, S. Y. et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral. Res. 181, 104873 (2020).
Liu, L. et al. Heparan sulfate proteoglycans as attachment factor for SARS-CoV-2. ACS Cent. Sci. 7, 1009–1018 (2021).
Hao, W. et al. Binding of the SARS-CoV-2 spike protein to glycans. Sci. Bull. 66, 1205–1214 (2021).
Thompson, A. J., de Vries, R. P. & Paulson, J. C. Virus recognition of glycan receptors. Curr. Opin. Virol. 34, 117–129 (2019).
Li, W. et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl Acad. Sci. USA 114, E8508–E8517 (2017).
Tortorici, M. A. et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 26, 481–489 (2019).
Weiss, R. J., Esko, J. D. & Tor, Y. Targeting heparin and heparan sulfate protein interactions. Org. Biomol. Chem. 15, 5656–5668 (2017).
Milewska, A. et al. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 88, 13221–13230 (2014).
Yang, Y., Du, Y. & Kaltashov, I. A. The utility of native MS for understanding the mechanism of action of repurposed therapeutics in COVID-19: heparin as a disruptor of the SARS-CoV-2 interaction with its host cell receptor. Anal. Chem. 92, 10930–10934 (2020).
Kwon, P. S. et al. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 6, 50 (2020).
Clausen, T. M. et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183, 1043–1057 (2020).
Morniroli, D., Gianni, M. L., Consales, A., Pietrasanta, C. & Mosca, F. Human sialome and coronavirus disease-2019 (COVID-19) pandemic: an understated correlation? Front. Immunol. 11, 1480 (2020).
Baker, A. N. et al. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 6, 2046–2052 (2020).
Ryzhikov, A. B. et al. Recombinant SARS-CoV-2 S protein binds to glycans of the lactosamine family in vitro. Biochemistry 86, 243–247 (2021).
Dhar, C. et al. Are sialic acids involved in COVID-19 pathogenesis? Glycobiology 31, 1068–1071 (2021).
Wu, S. C. et al. The SARS-CoV-2 receptor-binding domain preferentially recognizes blood group A. Blood Adv. 5, 1305–1309 (2021).
Yang, Q. et al. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. eLife 9, e61552 (2020).
Allen, J. D., Watanabe, Y., Chawla, H., Newby, M. L. & Crispin, M. Subtle influence of ACE2 glycan processing on SARS-CoV-2 recognition. J. Mol. Biol. 433, 166762 (2021).
El-Hawiet, A., Shoemaker, G. K., Daneshfar, R., Kitova, E. N. & Klassen, J. S. Applications of a catch and release electrospray ionization mass spectrometry assay for carbohydrate library screening. Anal. Chem. 84, 50–58 (2012).
Park, H. et al. Mass spectrometry-based shotgun glycomics for discovery of natural ligands of glycan-binding proteins. Anal. Chem. 92, 14012–14020 (2020).
Meloncelli, P. J. & Lowary, T. L. Synthesis of ABO histo-blood group type I and II antigens. Carbohydr. Res. 345, 2305–2322 (2010).
Meloncelli, P. J., West, L. J. & Lowary, T. L. Synthesis and NMR studies on the ABO histo-blood group antigens: synthesis of type III and IV structures and NMR characterization of type I-VI antigens. Carbohydr. Res. 346, 1406–1426 (2011).
Meloncelli, P. J. & Lowary, T. L. Synthesis of ABO histo-blood group type V and VI antigens. Austr. J. Chem. 62, 558–574 (2009).
Arungundram, S. et al. Modular synthesis of heparan sulfate oligosaccharides for structure-activity relationship studies. J. Am. Chem. Soc. 131, 17394–17405 (2009).
Elsaidi, H. R., Barreda, D. R., Cairo, C. W. & Lowary, T. L. Mycobacterial phenolic glycolipids with a simplified lipid aglycone modulate cytokine levels through Toll-like receptor 2. ChemBioChem 14, 2153–2159 (2013).
Sun, J., Kitova, E. N., Wang, W. & Klassen, J. S. Method for distinguishing specific from nonspecific protein-ligand complexes in nanoelectrospray ionization mass spectrometry. Anal. Chem. 78, 3010–3018 (2006).
Kitova, E. N., El-Hawiet, A., Schnier, P. D. & Klassen, J. S. Reliable determinations of protein–ligand interactions by direct ESI-MS measurements. Are we there yet? J. Am. Soc. Mass. Spectrom. 23, 431–441 (2012).
York, W. S. et al. GlyGen: computational and informatics resources for glycoscience. Glycobiology 30, 72–73 (2020).
Zhao, P. et al. Virus–receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe 28, 586–601 (2020).
Zhang, Y. et al. Mucin-type O-glycosylation landscapes of SARS-CoV-2 spike proteins. Preprint at https://doi.org/10.1101/2020.07.29.227785 (2020).
Shajahan, A., Supekar, N. T., Gleinich, A. S. & Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 30, 981–988 (2020).
Sanda, M., Morrison, L. & Goldman, R. N- and O-glycosylation of the SARS-CoV-2 spike protein. Anal. Chem. 93, 2003–2009 (2021).
Guile, G. R., Rudd, P. M., Wing, D. R., Prime, S. B. & Dwek, R. A. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal. Biochem. 240, 210–226 (1996).
Han, L. et al. Protein–glycolipid interactions studied in vitro using ESI-MS and nanodiscs: insights into the mechanisms and energetics of binding. Anal. Chem. 87, 4888–4896 (2015).
Bouwman, K. M. et al. Multimerization- and glycosylation-dependent receptor binding of SARS-CoV-2 spike proteins. PLoS Pathog. 17, e1009282 (2021).
Rillahan, C. D. et al. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat. Chem. Biol. 8, 661–668 (2012).
Albrecht, S. et al. Comprehensive profiling of glycosphingolipid glycans using a novel broad specificity endoglycoceramidase in a high-throughput workflow. Anal. Chem. 88, 4795–4802 (2016).
Stroh, L. J. et al. The greater affinity of JC polyomavirus capsid for α2,6-linked lactoseries tetrasaccharide c than for other sialylated glycans is a major determinant of infectivity. J. Virol. 89, 6364–6375 (2015).
Wu, W. & Air, G. M. Binding of influenza viruses to sialic acids: reassortant viruses with A/NWS/33 hemagglutinin bind to α2,8-linked sialic acid. Virology 325, 340–350 (2004).
Szklarczyk, O. M. et al. Receptor concentration and diffusivity control multivalent binding of Sv40 to membrane bilayers. PLoS Comput. Biol. 9, e1003310 (2013).
Song, E. et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 218, e20202135 (2021).
Sipione, S., Monyror, J., Galleguillos, D., Steinberg, N. & Kadam, V. Gangliosides in the brain: physiology, pathophysiology and therapeutic applications. Front. Neurosci. 14, 572965 (2020).
Salvatore, C. M. et al. Neonatal management and outcomes during the COVID-19 pandemic: an observation cohort study. Lancet Child Adolesc. Health 4, 721–727 (2020).
Li, J. et al. Association between ABO blood groups and risk of SARS-CoV-2 pneumonia. Br. J. Haematol. 190, 24–27 (2020).
Vitner, E. B. et al. Glucosylceramide synthase inhibitors prevent replication of SARS-CoV-2 and Influenza virus. J. Biol. Chem. 296, 100470 (2021).
Cho, J.-H. et al. Differential expression patterns of gangliosides in the tissues and cells of NIH-mini pig kidneys. Lab. Anim. Res. 14, 83–89 (2020).
Yang, T. J. et al. Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans. Proc. Natl Acad. Sci. USA 117, 1438–1446 (2020).
Stadlbauer, D. et al. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 57, e100 (2020).
Han, L. et al. Quantifying the binding stoichiometry and affinity of histo-blood group antigen oligosaccharides for human noroviruses. Glycobiology 28, 488–498 (2018).
Denisov, I. G., Grinkova, Y. V., Lazarides, A. A. & Sligar, S. G. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 (2004).
Kitov, P. I., Han, L., Kitova, E. N. & Klassen, J. S. Sliding window adduct removal method (SWARM) for enhanced electrospray ionization mass spectrometry binding data. J. Am. Soc. Mass. Spectrom. 30, 1446–1454 (2019).
Kitov, P. I. et al. A quantitative, high-throughput method identifies protein–glycan interactions via mass spectrometry. Commun. Biol. 2, 268 (2019).
Ceroni, A. et al. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome. Res. 7, 1650–1659 (2008).
Schmidt, F. et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 217, e20201181 (2020).
Schmitt, M. G., White, R. N. & Barnard, G. C. Development of a high cell density transient CHO plateform yielding mAb titers greater than 2g/L in only 7 days. Biotechnol. Prog. 36, e3047 (2020).
Yowler, B. C., Kensinger, R. D. & Schengrund, C. L. Botulinum neurotoxin A activity is dependent upon the presence of specific gangliosides in neuroblastoma cells expressing synaptotagmin I. J. Biol. Chem. 277, 32815–32819 (2002).
Acknowledgements
We acknowledge the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-06771 to J.S.K., RGPIN-2018-03815 to M.S.M.), ERC Starting Grant from the European Commission (802780 to R.P.dV), NIH National Heart, Lung and Blood Institute (R01HL151617 to G.-J.B.) the Canadian Glycomics Network, the Canada Foundation for Innovation and the Alberta Innovation and Advanced Education Research Capacity Program and the Beijerinck Premium of the Royal Dutch Academy of Sciences to R.P.dV. for generous funding. L.K.M. thanks the Canada Excellence Research Chair Program for funding. M.S.M. and L.J.W. thank Canada Research Chairs for a Chair in Chemical Glycoimmunology and Cardiac Transplantation, respectively. We thank D. J. Marchant (University of Alberta) for advice and critical reading the manuscript, J. Nagendran (University of Alberta) for generously providing the lung tissue, K. Susuki (University of Alberta) for preparation of lung homogenate, P. D. Bieniasz (The Rockefeller University) and J. L. M. Law (University of Alberta) for providing reagents (plasmids) for generating the pseudotyped virus and ACE2+ HEK293 cells, C. Cairo (University of Alberta) for the endoglycoceramidase enzyme and S. Sipione (University of Alberta) for advice related to glycolipids. mPlum-ER-3 was a gift from M. Davidson (Addgene plasmid 55966; http://n2t.net/addgene:55966; RRID:Addgene_55966). Plasmids for expression of SARS-CoV-2 S protein and RBD proteins were generously provided by F. Krammer (Icahn School of Medicine at Mount Sinai, produced under NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400008C) and produced under CEIRS contract HHSN272201400004C awarded to S.M.T. SARS-CoV-2 VIDO strain (hCoV-19/Canada/ON-VIDO-01/2020, GISAID accession EPI_ISL_425177) was kindly provided by D. Falzarano (Vaccine and Infectious Disease Organization). The SARS-CoV-2 72b Calg strain (72B/CA/CALG) was kindly provided by D. Evans (University of Alberta).
Author information
Authors and Affiliations
Contributions
J.S.K., M.S.M., R.P.dV., T.L.L., S.-T.D.H., L.K.M., T.H. and L.J.W. designed the research. L.N. and E.N.K. performed glycan screening, affinity measurements and data analysis. D.T.B. performed the N-glycan analysis. L.H. prepared the ganglioside NDs and performed screening. D.K. and G.C.D. synthesized MeαNeu5Ac and 3FNeu5Ac. K.A.M. performed the flow cytometry experiments examining RBD binding, performed pseudovirus infection and created knockout cell lines. K.M.B. and I.T. performed tissue staining with soluble RBD. M.E. performed the SARS-CoV-2 viral infection assays. S.M.T. contributed the SARS-CoV-2 RBD, and T.-J.Y. and S.-T.D.H. contributed the SARS-CoV-2 S protein. T.L.L. contributed the synthetic ABH blood group antigens and rhamnose-containing glycans. G.-J.B. and P.C. contributed HS. S.D.W. and A.L.M. prepared the intestinal tissue. K.A.M., L.N., M.S.M., R.P.dV. and J.S.K. wrote the paper. All authors contributed to review and editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemical Biology thanks Jeffrey Esko, Carlito Lebrilla and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Structures of 139 defined glycans used in CaR-ESI-MS screening against RBD and S protein.
These glycans comprise 9 groups: Lewis antigens, blood group ABH antigens, antigen-related glycans, globo-series glycans, ganglioside oligosaccharides, human milk oligosaccharides, Neu5Acα2-6 linked glycans, rhamnose-containing glycans (Library P) and others.
Extended Data Fig. 2 CaR-ESI-MS screening against defined glycan libraries at 25 °C.
Normalized abundances of released glycans from the SARS-CoV-2 RBD by CaR-ESI-MS at 25 °C. Summary of the charge-normalized (relative to 71) abundances of released ligands measured by CaR-ESI-MS screening of Library A – O against SARS-CoV-2 RBD. Measurements were performed in negative ion mode with a UHMR Orbitrap mass spectrometer at an HCD energy of 50 V. Aqueous ammonium acetate (100 mM, pH 6.9) solutions of SARS-CoV-2 RBD (10 μM) and glycan library (containing 50 nM of each glycan) at were used for screening. The different classes of oligosaccharides are distinguished by colour: red - Lewis antigens (1–14); yellow - blood group A antigens (15–20); orange - blood group B antigens (21–26); green - blood group H antigens (27–32); blue - sulfated compounds (34–38) and HS oligosaccharides (39–41), light blue - antigen-related glycans (42–50), purple - globo (51–60), pink - ganglioside oligosaccharides (61–76), white – HMOs and other glycans (77–128), light grey - Neu5Acα2-6-linked oligosaccharides (129–132), and dark grey – rhamnose-containing compounds (133–139). NA, not applicable. Asterisk indicates that the relative abundances (in CaR-ESI-MS) were estimated from their relative (to 69) Kd. Black-outlined white-filled circles represent data points. Data represent means ± s.d., n = 3 independent experiments.
Extended Data Fig. 3 Zero-charge mass spectrum for an aqueous ammonium acetate solution of SARS-CoV-2 RBD.
a, Zero-charge mass spectrum of RBD acquired in positive ion mode with a UHMR Orbitrap mass spectrometer for an aqueous ammonium acetate (100 mM, pH 6.9) solution of SARS-CoV-2 RBD (10 μM). Deconvolution was performed using the Thermo BioPharma Finder SoftwareTM. The glycan composition of each species is indicated (N ≡ HexNAc, H ≡ Hex, F ≡ Fuc, S ≡ Neu5Ac). b, Heatmap showing the distribution of the number of Neu5Ac and Fuc residues in the RBD glycoforms.
Extended Data Fig. 4 Comparison of glycan affinities for RBD measured by ESI-MS and their relative abundances measured by CaR-ESI-MS screening.
Comparison of glycan (15, 21, 27, 35, 36, 69, 71, 76 and 129) affinities for RBD measured by ESI-MS and their relative abundances measured by CaR-ESI-MS screening (performed at 25 °C). Black bars represent the ratio of the abundance of released glycans normalized to that of 71 under same conditions. White bars represent the ratios of dissociation affinities of RBD against 69 to dissociation affinities of RBD against glycan (L). ND, not detected. For relative abundances by CaR-ESI-MS, data represent ± s.d., n = 3 independent experiments. For Kd values, means ± s.d., n = 3 independent experiments for each glycan concentration.
Extended Data Fig. 5 CaR-ESI-MS screening ganglioside-containing nanodiscs against RBD.
a, ESI mass spectrum acquired in negative ion mode for an aqueous ammonium acetate solution (100 mM, 25 °C and pH 7.4) of RBD (10 µM) and 5 µM ganglioside-containing ND (1% GM1, GM2, GM3, GD1a, GD2, and GT1b, each at approximately 10 μM). b, CID mass spectrum acquired for ions centered at m/z 3,540, with a window width of 100 m/z units (the region highlighted in light blue in (a)) using a Trap collision energy of 50 V. c, CID mass spectrum acquired in negative ion mode for ions centered at m/z 3,540, with a window width of 100 m/z units, produced from an aqueous ammonium acetate solution (100 mM, 25 °C and pH 7.4) of and 10 µM ganglioside-containing ND; the Trap collision energy was 50 V.
Extended Data Fig. 6 ACE2 expression, RBD binding, and pseudoviral infection in HEK293 cells with and without ACE2.
a,b, Expression of human ACE2 (a) and trimeric fluorescent RBD binding (b) to WT (left) and ACE2+ (right) HEK293 cells as detected by flow cytometry. c, Flow cytometry gating of GFP-encoding pseudotyped SARS-CoV-2 lentivirus infection of WT (top) and ACE2+ (bottom) HEK293 cells.
Extended Data Fig. 7 Representative flow cytometry gating strategy and histogram for Fig. 4.
a-c, Side scatter (SSC) versus forward scatter (FSC) was used to discriminate against dead cells (SSC-A) and doublets (SSC-W). The singlets were then plotted in a histogram and the mean fluorescence intensity (MFI) was determined. Gating panels corresponding to Fig. 4a (a), 4b (b) and 4c (c). d, Representative gating strategy for Fig. 4d to determine % GFP+ cells in the pseudoviral infection assays when treated with 3FNeu5Ac. e, Representative flow cytometry gating and histogram for ACE2 binding in 3FNeu5Ac treated cells. This strategy corresponds to Fig. 4e.
Extended Data Fig. 8 Representative flow cytometry gating strategy and histogram for Fig. 5.
a, b, d, f, Side scatter (SSC) versus forward scatter (FSC) was used to discriminate against dead cells (SSC-A) and doublets (SSC-W). The singlets were then plotted in a histogram and the mean fluorescence intensity (MFI) was determined. Gating strategy of Genz-123346 treated cells (a, b) and UGCG+ or UGCG-/- HEK293 ACE2 cells (d, e, f) used to plot Fig. 5b(a), 5c (b), 5f (d), 5g (e), and 5h (f). c,g, Representative gating strategy for Fig. 5d(c) and 5i (g) to determine % GFP+ cells in the pseudoviral infection of HEK293 ACE2 UGCG+ and UGCG-/- cells.
Supplementary information
Supplementary Information
Supplementary Tables 1–4, Figs. 1–14 and Notes.
Supplementary Fig. 4
Statistical source data for Supplementary Fig. 4.
Supplementary Fig. 7
Statistical source data for Supplementary Fig. 7.
Source data
Source Data Fig. 1
Statistic source data.
Source Data Fig. 2
Statistic source data.
Source Data Fig. 3
Statistic source data.
Source Data Fig. 4
Statistic source data.
Source Data Fig. 5
Statistic source data.
Source Data Fig. 6
Statistic source data.
Rights and permissions
About this article
Cite this article
Nguyen, L., McCord, K.A., Bui, D.T. et al. Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2. Nat Chem Biol 18, 81–90 (2022). https://doi.org/10.1038/s41589-021-00924-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00924-1
This article is cited by
-
Sex drives colonic mucin sialylation in wild mice
Scientific Reports (2024)
-
Omicron Spike confers enhanced infectivity and interferon resistance to SARS-CoV-2 in human nasal tissue
Nature Communications (2024)
-
The role of galectins in immunity and infection
Nature Reviews Immunology (2023)
-
Metabolic alterations upon SARS-CoV-2 infection and potential therapeutic targets against coronavirus infection
Signal Transduction and Targeted Therapy (2023)
-
Role of ST6GAL1 and ST6GAL2 in subversion of cellular signaling during enteroaggregative Escherichia coli infection of human intestinal epithelial cell lines
Applied Microbiology and Biotechnology (2023)