Abstract
Spindle position control is essential for cell fate determination and organogenesis. Early studies indicate the essential role of the evolutionarily conserved Gαi/LGN/NuMA network in spindle positioning. However, the regulatory mechanisms that couple astral microtubules dynamics to the spindle orientation remain elusive. Here we delineated a new mitosis-specific crotonylation-regulated astral microtubule–EB1–NuMA interaction in mitosis. EB1 is a substrate of TIP60, and TIP60-dependent crotonylation of EB1 tunes accurate spindle positioning in mitosis. Mechanistically, TIP60 crotonylation of EB1 at Lys66 forms a dynamic link between accurate attachment of astral microtubules to the lateral cell cortex defined by NuMA–LGN and fine tune of spindle positioning. Real-time imaging of chromosome movements in HeLa cells expressing genetically encoded crotonylated EB1 revealed the importance of crotonylation dynamics for accurate control of spindle orientation during metaphase–anaphase transition. These findings delineate a general signaling cascade that integrates protein crotonylation with accurate spindle positioning for chromosome stability in mitosis.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting our findings in this study are available in this paper and the Supplementary Data. The structure of EB3–microtubule (PDB entry 3JAK) and EB1CH domain (PDB 1PA7) were from PDB. Source data are provided with this paper.
References
Cheeseman, I. M. & Desai, A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 (2008).
Yao, X. B., Abrieu, A., Zheng, Y., Sullivan, K. F. & Cleveland, D. W. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491 (2000).
Pearson, C. G. & Bloom, K. Dynamic microtubules lead the way for spindle positioning. Nat. Rev. Mol. Cell Biol. 5, 481–492 (2004).
Thery, M. et al. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 7, 947–953 (2005).
Hendricks, A. G. et al. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr. Biol. 22, 632–637 (2012).
Seldin, L., Muroyama, A. & Lechler, T. NuMA-microtubule interactions are critical for spindle orientation and the morphogenesis of diverse epidermal structures. eLife 5, e12504 (2016).
Kiyomitsu, T. & Cheeseman, I. M. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14, 311–317 (2012).
Akhmanova, A. & Steinmetz, M. O. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 (2008).
Jiang, K. et al. TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 10, 857–865 (2009).
Lu, M. S. & Johnston, C. A. Molecular pathways regulating mitotic spindle orientation in animal cells. Development 140, 1843–1856 (2013).
Hayashi, I. & Ikura, M. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J. Biol. Chem. 278, 36430–36434 (2003).
Xia, P. et al. EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl Acad. Sci. USA 109, 16564–16569 (2012).
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Wan, J., Liu, H., Chu, J. & Zhang, H. Functions and mechanisms of lysine crotonylation. J. Cell. Mol. Med. 23, 7163–7169 (2019).
Mo, F. et al. Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat. Chem. Biol. 12, 226–232 (2016).
Wu, Q. et al. Ultradeep lysine crotonylome reveals the crotonylation enhancement on both histones and nonhistone proteins by SAHA treatment. J. Proteome Res. 16, 3664–3671 (2017).
Kim, C. H., Kang, M., Kim, H. J., Chatterjee, A. & Schultz, P. G. Site-specific incorporation of epsilon-N-crotonyllysine into histones. Angew. Chem. Int. Ed. Engl. 51, 7246–7249 (2012).
Qin, X. et al. An orthogonal tyrosyl-tRNA synthetase/tRNA pair from a thermophilic bacterium for an expanded eukaryotic genetic code. Biochemistry 59, 90–99 (2020).
McKinley, K. L. & Cheeseman, I. M. Large-scale analysis of CRISPR/Cas9 cell-cycle knockouts reveals the diversity of p53-dependent responses to cell-cycle defects. Dev. Cell 40, 405–420 e2 (2017).
Wei, W. et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 27, 898–915 (2017).
Ishii, S., Kurasawa, Y., Wong, J. & Yu-Lee, L. Y. Histone deacetylase 3 localizes to the mitotic spindle and is required for kinetochore-microtubule attachment. Proc. Natl Acad. Sci. USA 105, 4179–4184 (2008).
Lahm, A. et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl Acad. Sci. USA 104, 17335–17340 (2007).
Bao, X. et al. Mitosis-specific acetylation tunes Ran effector binding for chromosome segregation. J. Mol. Cell. Biol. 10, 18–32 (2018).
Zhao, G. et al. Dynamic acetylation of the kinetochore-associated protein HEC1 ensures accurate microtubule-kinetochore attachment. J. Biol. Chem. 294, 576–592 (2019).
Huang, Y. et al. BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules. Cell Res. 29, 562–578 (2019).
Yu, H. et al. NDP52 tunes cortical actin interaction with astral microtubules for accurate spindle orientation. Cell Res. 29, 666–679 (2019).
di Pietro, F., Echard, A. & Morin, X. Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 17, 1106–1130 (2016).
Zhang, R., Alushin, G. M., Brown, A. & Nogales, E. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell 162, 849–859 (2015).
van Haren, J. et al. Local control of intracellular microtubule dynamics by EB1 photodissociation. Nat. Cell Biol. 20, 252–261 (2018).
Aher, A. et al. CLASP suppresses microtubule catastrophes through a single TOG domain. Dev. Cell 46, 40–58 e8 (2018).
Morin, X. & Bellaiche, Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell 21, 102–119 (2011).
Jiang, K. et al. A proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22, 1800–1807 (2012).
Yao, X. & Smolka, A. J. Gastric parietal cell physiology and Helicobacter pylori-induced disease. Gastroenterology 156, 2158–2173 (2019).
Liu, X. et al. Mitotic motor CENP-E cooperates with PRC1 in temporal control of central spindle assembly. J. Mol. Cell. Biol. 12, 654–665 (2020).
Pirovano, L. et al. Hexameric NuMA:LGN structures promote multivalent interactions required for planar epithelial divisions. Nat. Commun. 10, 2208 (2019).
Yang, Y. et al. CYLD regulates spindle orientation by stabilizing astral microtubules and promoting dishevelled-NuMA-dynein/dynactin complex formation. Proc. Natl Acad. Sci. USA 111, 2158–2163 (2014).
Mora-Bermudez, F., Matsuzaki, F. & Huttner, W. B. Specific polar subpopulations of astral microtubules control spindle orientation and symmetric neural stem cell division. eLife 3, e02875 (2014).
Toyoshima, F. & Nishida, E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26, 1487–1498 (2007).
Bouissou, A. et al. gamma-tubulin ring complexes and EB1 play antagonistic roles in microtubule dynamics and spindle positioning. EMBO J. 33, 114–128 (2014).
Gallini, S. et al. NuMA phosphorylation by Aurora-A orchestrates spindle orientation. Curr. Biol. 26, 458–469 (2016).
Kotak, S., Afshar, K., Busso, C. & Gonczy, P. Aurora A kinase regulates proper spindle positioning in C. elegans and in human cells. J. Cell Sci. 129, 3015–3025 (2016).
Polverino, F. et al. The Aurora-A/TPX2 Axis directs spindle orientation in adherent human cells by regulating NuMA and microtubule stability. Curr. Biol. 31, 658–667.e5 (2020).
Du, Q., Taylor, L., Compton, D. A. & Macara, I. G. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr. Biol. 12, 1928–1933 (2002).
Li, X. et al. Structure-guided development of YEATS domain inhibitors by targeting pi-pi-pi stacking. Nat. Chem. Biol. 14, 1140–1149 (2018).
Yao, X. B., Anderson, K. L. & Cleveland, D. W. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435–447 (1997).
Xia, P. et al. Superresolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol. Biol. Cell 25, 4166–4173 (2014).
Song, X. et al. Acetylation of ACAP4 regulates CCL18-elicited breast cancer cell migration and invasion. J. Mol. Cell. Biol. 10, 559–572 (2018).
Song, X. et al. Acetylation of ezrin regulates membrane-cytoskeleton interaction underlying CCL18-elicited cell migration. J. Mol. Cell. Biol. 12, 424–437 (2020).
Forester, T. R. & Smith, W. SHAKE, rattle, and roll: efficient constraint algorithms for linked rigid bodies. J. Comput. Chem. 19, 102–111 (1998).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald—an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Wang, T. et al. SENP1-Sirt3 signaling controls mitochondrial protein acetylation and metabolism. Mol. Cell 75, 823–834 e5 (2019).
Acknowledgements
We are grateful to Y. Shi, Y. Chen, Y. Luo, H. Jiang, S. Harris-Hooker and G. Tosini for support, and P. Schultz and I.M. Cheeseman for reagents. This work was supported by the MOST-NSFC grants (nos. 2017YFA0503600, 91854203, 31621002, 91853115, 21922706, 31671405, 32090040 and 2016YFA0100500 to Xing L.; 2016YFA0101202, 31970655 to Z.D., 91953101 to Z.Z. and 92059102 to X.S.); the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB19040000) to Xing L.; the Ministry of Education (grant nos. IRT_17R102, 20113402130010; YD2070006001 to Xing L.); the Fundamental Research Funds for the Central Universities (grant no. WK2070000194 to Xing L.); Anhui Provincial Natural Science Foundation grant (no. 1908085MC64); and a China Postdoctoral Science Foundation grant (no. 2019M662184) to X.S. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
X.Y. and Xing L. conceived the project. X.S., F.Y. and P.X. designed and performed most of the biochemical and cell biological experiments. Y.W., X.S. and K.J. performed EB1 TIRF experiments in vitro. Z.Z. performed molecular dynamics simulations. Xu L., and X.Y. designed and performed the organoids experiments. Z.W. and M.M. performed the chemical biological experiments and evaluated small molecule inhibitors. X.Y. and R.T. performed MS identification. R.Z. and K.R. performed structure modeling and labeling. Z.D. and T.L. assisted in recombinant protein engineering and purification. L.L, J.L., C.X., W.Y., B.H., J.Z. and J.C. contributed reagents. X.S., P.X., F.Y. and Xu L. performed data analyses. X.S., Xing L. and X.Y. wrote the paper. D.L.H. edited the paper. All authors have commented on and approved the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 EB1 interacts with and is crotonylated by TIP60.
(a) HEK293T cells co-transfected with FLAG-EB1 and GFP, GFP-PCAF, GFP-TIP60 or myc-p300. 24 hours after transfection, FLAG-EB1 was immuno-purified and subject to Western blotting analyses, and crotonylation of EB1 was judged by anti-pan-CrK antibody. (b) In vitro TIP60 acetylation/crotonylation reactions were performed with crotonyl-CoA (Cr-CoA) and acetyl-CoA (Ac-CoA). Reaction products were immunoblotted with the indicated antibodies. FLAG-TIP60 was purified from HEK293T cells. Aurora B-mCherry-His and p53-mCherry-His were purified from E. coli and used as substrates. The acetylation and crotonylation level were detected with an anti-pan-AcK and anti-pan-CrK antibody. Protein levels were analyzed with SDS-PAGE gel stained by CBB. (c) HEK293T cells were co-transfected with FLAG-EB1 and GFP or GFP-TIP60 for 24 h, and their interactions were tested by co-immunoprecipitation. Western blotting analyses were conducted by anti-GFP and FLAG. (d) HEK293T cells were co-transfected with FLAG-TIP60 and GFP or EB1-GFP for 24 h, and their interactions were tested by co-immunoprecipitation. Western blotting analyses were conducted by anti-GFP and FLAG antibodies. (e) Schematic illustration of EB1 structural domains and truncation mutants. Their binding properties with TIP60 were also annotated. (f) Bacterially expressed GST-EB1 full-length (FL) and truncation mutants were purified and used as affinity matrices to isolate His-TIP60. The binding fraction was analyzed by immunoblotting and protein levels were analyzed with SDS-PAGE gel stained by CBB. An asterisk indicates the band corresponding to the protein of interest, if multiple bands were observed. (g) Schematic illustration of TIP60 structural domains and truncation mutants. (h) Bacterially expressed GST-EB1 was purified and used as affinity matrix to isolate GFP-TIP60 full-length (FL) or truncation mutants expressed in HEK293T cells. Binding activity was analyzed by immunoblotting with an anti-GFP antibody. (i) The FLAG-EB1 proteins from Fig. 1a were analyzed by mass spectrometry. The typical EB1 Lys66 crotonylation mass spectrum was shown. Related to Fig. 1.
Extended Data Fig. 2 The generation of CrK66- and AcK66-EB1 proteins by unnatural amino acids (UAAs) incorporation method in E.coli.
(a) Schematic diagram showing the expression of CrK66-EB1 and AcK66-EB1 in E. coli. The UAG in the mRNA of EB1 was decoded by tRNACUA in the presence of N-ε-crotonyl-lysine or N-ε-acetyl-lysine, giving rise to CrK66-EB1 or AcK66-EB1. NAM (10 mM) was added to prevent decrotonylation or deacetylation. (b) Diagram of the plasmid construction used to express recombinant CrK66/AcK66 and wild-type EB1 in E.coli. (c) Validation of AcK66-EB1 and CrK66-EB1 proteins generated as in b via CBB staining. Purified wild-type, AcK66- and CrK66-EB1 were also analyzed by AcK66-EB1 and CrK66-EB1 antibodies. (d) The specificity of CrK66-EB1 and AcK66-EB1 antibodies and the EB1 knockout efficiency were detected in CRISPR/Cas9-mediated EB1 knockout HeLa cells. Doxycycline (Dox, 1 μg/mL) was added to induce EB1 knockout for indicated time. Cell lysates were probed with anti-CrK66-EB1, AcK66-EB1 and EB1 antibodies. (e) HeLa cells depleted of endogenous EB1 were fixed and stained with CrK66-EB1, EB1 and α-tubulin antibodies. DNA was stained with DAPI. Scale bar, 10 μm. (f) HeLa cells were synchronized to the indicated time points by double thymidine release. Whole cell lysates (WCL) were probed for Lys66 crotonylation and acetylation levels by Western blotting with indicated antibodies. (g-h) Quantification of CrK66-EB1 and AcK66-EB1 levels in synchronized HeLa cells at G1/S or mitosis. Purified CrK66-EB1 and AcK66-EB1 proteins from E. coli were used as standard. Related to Fig. 1.
Extended Data Fig. 3 HDAC3 interacts with and decrotonylates EB1.
(a) FLAG-HDAC1-3 or FLAG-SIRT1-3 were co-transfected with EB1-GFP in HEK293T cells, and their interactions were tested by co-immunoprecipitation. (b) Whole-cell Lysates (WCL) of HeLa cells expressing FLAG-HDAC3 wild type or deacetylase-dead point mutants (Y298H and VRPP) were probed with indicated antibodies, including site-specific modification of antibodies. (c) Whole-cell Lysates (WCL) of HeLa cells expressing HDAC3 shRNA or FLAG-HDAC3 were probed with an anti-CrK66 antibody to assess the crotonylation level of EB1. (d) FLAG-TIP60 was incubated with His-EB1 in the presence of indicated concentrations of Cr-CoA or Ac-CoA for in vitro acylation assay. The acetylation and crotonylation levels of EB1 were analyzed with anti-AcK66-EB1 and CrK66-EB1 antibodies. Purified proteins were validated via CBB staining. Related to Fig. 1.
Extended Data Fig. 4 TIP60 inhibition causes spindle orientation defects in mitotic cells.
(a) Schematic illustration of NU9056, a TIP60 inhibitor, treatment in HeLa cells at metaphase-anaphase transition. (b) Representative phenotypes of metaphase-anaphase transition in HeLa cells treated with DMSO or NU9056 (20 μM). Scale bar, 10 μm. (c) Statistical analyses of time from metaphase to anaphase onset in cells treated as in b. DMSO, n = 25; NU9056, n = 25. Data represent mean ± s.e.m. from three independent experiments. Statistical significance was determined by two-sided t-test (***p < 0.0001). (d) Quantification of mitotic phenotypes in HeLa cells treated as in b. DMSO, n = 25; NU9056, n = 25. Data represent mean ± s.e.m. (e) MG132-released HeLa cells were treated with DMSO or NU9056 (20 μM) for 1 h before harvest. Whole-cell lysates (WCL) were separated by SDS-PAGE and blotted with the indicated antibodies. (f) HeLa cells were transfected with FLAG-Hec1 and arrested at metaphase by MG132. MG132-released HeLa cells were then treated with DMSO or NU9056 for 1 h followed by immunoprecipitation with FLAG resin. Immunoisolated Hec1 were subjected to Western blotting with an anti-acetyllysine antibody (AcK) to analyze the acetylation level. Related to Fig. 2.
Extended Data Fig. 5 The generation of CrK66- and AcK66-EB1 proteins by UAAs incorporation method in HeLa cells.
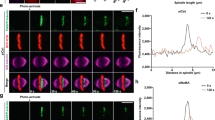
(a) Diagram of the plasmid construction used to express recombinant CrK66 or wild-type EB1 in HeLa cells depleted of endogenous EB1. (b) EB1-WT-GFP or EB1-K66TAG-GFP and pCMV-CrKRS/ acKRS were co-transfected into HeLa cells depleted endogenous EB1. CRISPR/Cas9-mediated endogenous EB1 knockout was achieved by addition of Doxycycline (Dox, 1 μg/mL) into CRISPR/Cas9-edited HeLa cells. The expression levels of EB1-GFP and CrK66-EB1 or AcK66-EB1 were analyzed by Western blotting with anti-GFP, anti-AcK66-EB1 and anti-CrK66-EB1 antibodies. (c) Schematic illustration of spindle positioning in mitotic HeLa cells expressing wild-type or Lys66-crotonylated EB1. Note that EB1 crotonylation at Lys66 causes slope of spindle in the z direction, which means that, when one spindle pole is just right on the focal plane, the other pole usually stays out of focus. Related to Fig. 2.
Extended Data Fig. 6 EB1 Lys66 crotonylation regulates microtubule stability.
(a) Multi-alignment of EB1 from different species, and the conserved K66 site of EB1 was labeled by red triangle. (b) Overall structure of Fig. 3a. (c) Cartoon drawing of in vitro microtubule plus-end-tracking assay for EB1 using TIRFM. (d) Quantification of contact number of β-tubulin and EB1 wild type and mutants in 0.1 ns. For each group, n = 1,000. Data represent mean ± s.e.m. from three independent experiments. Ordinary one-way ANOVA followed by Tukey’s post hoc test was used to determine statistical significance (***p < 0.0001). (e) The contacts number revolution in All-atom molecular dynamics simulations of EB1 wild type and mutants for 100 ns. (f) The last conformation of All-atom molecular dynamics simulation in EB1 wild type and mutants. The conformation is colored by light blue (α-tubulin), pale green (β-tubulin) and orange (EB1). Lys66 (light pink) and Glu386 (green) are showed by CPK model. Related to Fig. 3 & Supplementary table 1.
Extended Data Fig. 7 EB1 crotonylation regulates astral microtubule stability in cells.
(a) EB1-WT-GFP or EB1-K66TAG-GFP and pCMV-CrKRS/ acKRS were co-transfected into HeLa cells depleted of endogenous EB1. CRISPR/Cas9-mediated endogenous EB1 knockout was achieved by addition of Doxycycline (Dox, 1 μg/mL) into CRISPR/Cas9-edited HeLa cells. Cells were fixed by precooling methanol and stained with anti-γ-tubulin antibody. Scale bar, 10 μm. (b) Endogenous EB1-depleted HeLa cells expressing EB1-WT-GFP, AcK66-EB1-GFP and CrK66-EB1-GFP with or without FLAG-EB1CH-TOG2, and the acylation level and FLAG-EB1CH-TOG2 expression level were analyzed by indicated antibodies. (c) HeLa cells expressing NDP52 siRNA were transfected with or without FLAG-EB1CH-TOG2. Cells were synchronized to metaphase with MG132 treatment, fixed in pre-warmed paraformaldehyde and stained with α-tubulin antibody. The boxed areas in the α-tubulin channel are magnified in the right panels to show details of the astral microtubule region. Scale bar, 10 μm. (d) HeLa cells were transfected with NDP52 siRNA for 48 h. The knockdown efficiency of NDP52 siRNA and FLAG-EB1CH-TOG2 expression level were assessed by Western blotting. (e-f) Statistical analysis of astral microtubule number (e) and intensity (f) in c. For astral microtubule number, only microtubule length above 2 μm was included in statistics, for astral microtubule intensity, the average intensity was achieved by total intensity dividing the astral microtubule area. Control siRNA, n = 20; NDP52 siRNA, n = 20; NDP52 siRNA + TOG2, n = 20. Data represent mean ± s.e.m. from three independent experiments. Ordinary one-way ANOVA followed by Tukey’s post hoc test was used to determine statistical significance. NS (not significant) indicates p > 0.05. Related to Fig. 4.
Extended Data Fig. 8 The interaction between EB1 and NuMA is mediated by EEY motif.
(a) EB1 binding of purified + TIP fragments in vitro. Recombinant + TIP proteins were purified and incubated with recombinant wild-type, AcK66- and CrK66-EB1 for pull-down assay. Their interaction was assessed by Western blotting with an anti-EB1 antibody. (b) Representative immunofluorescence images of endogenous EB1-depleted HeLa cells expressing EB1-WT-GFP and CrK66-EB1-GFP. Cells were fixed and stained with NuMA and α-tubulin antibodies. The boxed areas are magnified in the right panels to show details of the astral microtubule region. Scale bar, 10 μm. For the whole-cell images were shown as Z-stacks, while zoom-in images as single-plane images. Arrowheads indicate the cortex NuMA in whole-cell images, and arrowheads indicate EB1 dots that co-localize with cortex NuMA. (c) Endogenous EB1-depleted HeLa cells expressing EB1-WT-GFP or CrK66-EB1-GFP, AcK66-EB1-GFP with mCherry-LGN. The whole-cell and kymograph images of indicated region were shown. Arrowheads indicate EB1 near to the cortex. (d) In vitro pull-down assay. Recombinant EB1, AcK66-EB1 and CrK66-EB1 were captured on Ni-NTA resin followed by incubation with mitotic HeLa cell lysates. The binding fractions were analyzed by Western blotting and mass spectrometry. (e) Sequence alignment of NuMA homologs from different species. Two conserved SxIP motifs in NuMA-CT were annotated with blue pentastar. (f) Bacterially expressed GST-NuMA-CT was purified and used as affinity matrix to isolate His-EB1-WT and CrK66. Binding activity was analyzed with an anti-EB1 antibody and protein levels were shown by staining with CBB. (g) Recombinant GST-NuMA-CT and SxIP-motif mutants were incubated with His-EB1 for GST pull-down assay. NN#1, NN#2 and NNNN were the asparagine-substituted mutants of the SxIP motifs in e. (h) Recombinant GST-NuMA-CT and its deletion mutants were incubated with His-EB1 for GST pull-down assay. Their interaction was assessed by Western blotting with an anti-EB1 antibody and protein levels were shown by staining with CBB. Related to Fig. 5 & Supplementary Table 2.
Extended Data Fig. 9 Increasing cellular crotonylation level compromises EB1 localization to mitotic spindle in mouse gastric organoids.
(a) Cartoon presentation of generation of mouse-derived gastric organoids. The organoid model provides a unique platform to delineate cellular dynamics during cell division in native tissue. (b) Representative immunofluorescence images of EB1 localization in mitotic cells in gastric organoids derived from wild-type mice. Gastric organoids were fixed and stained with EB1 (red), DM1A (green), and DAPI (blue). Scale bar, 10 μm. (c) Representative immunofluorescence images of EB1 localization in mitotic cells in gastric organoids derived from wild-type and TIP60-KI mice. Scale bar, 10 μm. (d) Statistical analyses of EB1 fluorescence intensity in mitotic spindle in c. Wild-type, n = 24; TIP60 KI, n = 24. Data represent mean ± s.e.m. from three independent experiments. Statistical significance was determined by two-sided t-test (***p < 0.0001). Related to Fig. 6.
Supplementary information
Supplementary Table 1
Microtubule growth rate and catastrophe frequency were determined from kymograph images of Fig. 3b.
Supplementary Table 2
MS data of Extended Data Fig. 8d.
Supplementary Data
Statistical source data of Supplementary Table 1.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Western blots and/or gels.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Western blots and/or gels.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Western blots and/or gels.
Source Data Extended Data Fig. 1
Western blots and/or gels.
Source Data Extended Data Fig. 2
Western blots and/or gels.
Source Data Extended Data Fig. 3
Western blots and/or gels.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Western blots and/or gels.
Source Data Extended Data Fig. 5
Western blots and/or gels.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Western blots and/or gels.
Source Data Extended Data Fig. 8
Western blots and/or gels.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Song, X., Yang, F., Liu, X. et al. Dynamic crotonylation of EB1 by TIP60 ensures accurate spindle positioning in mitosis. Nat Chem Biol 17, 1314–1323 (2021). https://doi.org/10.1038/s41589-021-00875-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00875-7
This article is cited by
-
The two sides of chromosomal instability: drivers and brakes in cancer
Signal Transduction and Targeted Therapy (2024)
-
Spatiotemporal and direct capturing global substrates of lysine-modifying enzymes in living cells
Nature Communications (2024)
-
A glimpse into novel acylations and their emerging role in regulating cancer metastasis
Cellular and Molecular Life Sciences (2024)
-
SEPT2 crotonylation promotes metastasis and recurrence in hepatocellular carcinoma and is associated with poor survival
Cell & Bioscience (2023)
-
Phase separation of EB1 guides microtubule plus-end dynamics
Nature Cell Biology (2023)