Abstract
Polycomb repressive complex 1 (PRC1) is an essential chromatin-modifying complex that monoubiquitinates histone H2A and is involved in maintaining the repressed chromatin state. Emerging evidence suggests PRC1 activity in various cancers, rationalizing the need for small-molecule inhibitors with well-defined mechanisms of action. Here, we describe the development of compounds that directly bind to RING1B–BMI1, the heterodimeric complex constituting the E3 ligase activity of PRC1. These compounds block the association of RING1B–BMI1 with chromatin and inhibit H2A ubiquitination. Structural studies demonstrate that these inhibitors bind to RING1B by inducing the formation of a hydrophobic pocket in the RING domain. Our PRC1 inhibitor, RB-3, decreases the global level of H2A ubiquitination and induces differentiation in leukemia cell lines and primary acute myeloid leukemia (AML) samples. In summary, we demonstrate that targeting the PRC1 RING domain with small molecules is feasible, and RB-3 represents a valuable chemical tool to study PRC1 biology.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq and ChIP–seq data for TEX cells treated with RB-3 have been submitted to the Gene Expression Omnibus (GEO) database under accession codes GSE123490 and GSE123930, respectively. Structures of RING1B–BMI1f and RING1B–BMI1f cocrystalized in the presence of RB-2 were deposited in the PDB under the accession codes 6WI7 and 6WI8, respectively. The structure of the RING1B–BMI1f–RB-2 complex determined using a hybrid approach was deposited in the PDB under the accession code 7ND1. PDB structures 3RPB and 4R8P were used for data analysis. Source data are provided with this paper.
References
Simon, J. A. & Kingston, R. E. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 (2009).
Chittock, E. C., Latwiel, S., Miller, T. C. & Muller, C. W. Molecular architecture of Polycomb repressive complexes. Biochem. Soc. Trans. 45, 193–205 (2017).
Gao, Z. et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 (2012).
Taherbhoy, A. M., Huang, O. W. & Cochran, A. G. BMI1–RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun. 6, 7621 (2015).
Scelfo, A. et al. Functional landscape of PCGF proteins reveals both RING1A/B-dependent- and RING1A/B-independent-specific activities. Mol. Cell 74, 1037–1052 (2019).
Buchwald, G. et al. Structure and E3-ligase activity of the Ring–Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 (2006).
Tamburri, S. et al. Histone H2AK119 mono-ubiquitination is essential for Polycomb-mediated transcriptional repression. Mol. Cell 77, 840–856 (2020).
Blackledge, N. P. et al. PRC1 catalytic activity is central to Polycomb system function. Mol. Cell 77, 857–874 (2020).
McGinty, R. K., Henrici, R. C. & Tan, S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014).
Blackledge, N. P. et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and Polycomb domain formation. Cell 157, 1445–1459 (2014).
Rose, N. R. et al. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. eLife 5, e18591 (2016).
Fursova, N. A. et al. Synergy between variant PRC1 complexes defines Polycomb-mediated gene repression. Mol. Cell 74, 1020–1036 (2019).
Bonnet, D. & Dick, J. E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 (1997).
Kreso, A. et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med. 20, 29–36 (2014).
Martin-Perez, D., Piris, M. A. & Sanchez-Beato, M. Polycomb proteins in hematologic malignancies. Blood 116, 5465–5475 (2010).
Sauvageau, M. & Sauvageau, G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7, 299–313 (2010).
Lessard, J. & Sauvageau, G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255–260 (2003).
Shima, H. et al. Ring1A and Ring1B inhibit expression of Glis2 to maintain murine MOZ–TIF2 AML stem cells. Blood 131, 1833–1845 (2018).
Rizo, A. et al. Repression of BMI1 in normal and leukemic human CD34+ cells impairs self-renewal and induces apoptosis. Blood 114, 1498–1505 (2009).
van den Boom, V. et al. Non-canonical PRC1.1 targets active genes independent of H3K27me3 and is essential for leukemogenesis. Cell Rep. 14, 332–346 (2016).
Alchanati, I. et al. The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2α cleavage complex—a potentially new drug target. PLoS ONE 4, e8104 (2009).
Su, W. et al. The Polycomb repressor complex 1 drives double-negative prostate cancer metastasis by coordinating stemness and immune suppression. Cancer Cell 36, 139–155 (2019).
Li, Z. et al. Structure of a Bmi-1–Ring1B Polycomb group ubiquitin ligase complex. J. Biol. Chem. 281, 20643–20649 (2006).
van Zundert, G. C. P. et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016).
Bhatnagar, S. et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature 516, 116–120 (2014).
Kalb, R., Mallery, D. L., Larkin, C., Huang, J. T. & Hiom, K. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 8, 999–1005 (2014).
Horn, V. et al. Structural basis of specific H2A K13/K15 ubiquitination by RNF168. Nat. Commun. 10, 1751 (2019).
Dixon, A. S. et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 (2016).
Warner, J. K. et al. Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia 19, 1794–1805 (2005).
McDermott, S. P. et al. A small molecule screening strategy with validation on human leukemia stem cells uncovers the therapeutic efficacy of kinetin riboside. Blood 119, 1200–1207 (2012).
van Rhenen, A. et al. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin. Cancer Res. 11, 6520–6527 (2005).
Pulikkan, J. A., Tenen, D. G. & Behre, G. C/EBPα deregulation as a paradigm for leukemogenesis. Leukemia 31, 2279–2285 (2017).
Yu, M. et al. Direct recruitment of Polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol. Cell 45, 330–343 (2012).
Barbour, H., Daou, S., Hendzel, M. & Affar, E. B. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat. Commun. 11, 5947 (2020).
Cales, C. et al. Inactivation of the Polycomb group protein Ring1B unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with Ink4a deletion. Mol. Cell. Biol. 28, 1018–1028 (2008).
Nishida, Y. et al. Preclinical activity of the novel B-cell-specific Moloney murine leukemia virus integration site 1 inhibitor PTC-209 in acute myeloid leukemia: implications for leukemia therapy. Cancer Sci. 106, 1705–1713 (2015).
Barabe, F., Kennedy, J. A., Hope, K. J. & Dick, J. E. Modeling the initiation and progression of human acute leukemia in mice. Science 316, 600–604 (2007).
Jeusset, L. M. & McManus, K. J. Developing targeted therapies that exploit aberrant histone ubiquitination in cancer. Cells 8, 165 (2019).
Park, I. K. et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305 (2003).
Su, W. J. et al. RNF2/Ring1b negatively regulates p53 expression in selective cancer cell types to promote tumor development. Proc. Natl Acad. Sci. USA 110, 1720–1725 (2013).
Ardley, H. C. Ring finger ubiquitin protein ligases and their implication to the pathogenesis of human diseases. Curr. Pharm. Des. 15, 3697–3715 (2009).
Duffy, M.J., Synnott, N.C., O’Grady, S. & Crown, J. Targeting p53 for the treatment of cancer. Semin. Cancer Biol. https://doi.org/10.1016/j.semcancer.2020.07.005 (2020).
Watt, G. F., Scott-Stevens, P. & Gaohua, L. Targeted protein degradation in vivo with proteolysis targeting chimeras: current status and future considerations. Drug Discov. Today Technol. 31, 69–80 (2019).
Gray, F. et al. BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization. Nat. Commun. 7, 13343 (2016).
Jaatinen, T. et al. Global gene expression profile of human cord blood-derived CD133+ cells. Stem Cells 24, 631–641 (2006).
Gal, H. et al. Gene expression profiles of AML derived stem cells; similarity to hematopoietic stem cells. Leukemia 20, 2147–2154 (2006).
Sheffield, P. J., Derewenda, U., Taylor, J., Parsons, T. J. & Derewenda, Z. S. Expression, purification and crystallization of a BH domain from the GTPase regulatory protein associated with focal adhesion kinase. Acta Crystallogr. D Biol. Crystallogr. 55, 356–359 (1999).
Brzovic, P. S. et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl Acad. Sci. USA 100, 5646–5651 (2003).
Wiener, R. et al. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 20, 1033–1039 (2013).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Tugarinov, V. & Kay, L. E. Quantitative NMR studies of high molecular weight proteins: application to domain orientation and ligand binding in the 723 residue enzyme malate synthase G. J. Mol. Biol. 327, 1121–1133 (2003).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Tugarinov, V. & Kay, L. E. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 125, 13868–13878 (2003).
Huang, H. et al. Covalent inhibition of NSD1 histone methyltransferase. Nat. Chem. Biol. 16, 1403–1410 (2020).
Egan, B. et al. An alternative approach to ChIP–seq normalization enables detection of genome-wide changes in histone H3 lysine 27 trimethylation upon EZH2 inhibition. PLoS ONE 11, e0166438 (2016).
Grembecka, J. et al. Menin–MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 8, 277–284 (2012).
Acknowledgements
This work was funded by the National Institute of Health (NIH) R01 grants CA207272, CA226759 and CA240514 to T.C., CA201204, CA244254 and CA160467 to J.G. and LLS Scholar grants (1340-17) to T.C. and (1215-14) to J.G. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract number DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). TEX and MLL–ENL cells were received from J. Dick, University Health Network, Toronto, Canada. We thank M. Carroll and G. Danet-Desnoyers from the Stem Cell and Xenograft Core at the University of Pennsylvania for providing a human AML primary sample.
Author information
Authors and Affiliations
Contributions
T.C. and J.G. were responsible for initiating and supervising the project. S.S., F.G., H.M., P.G.-A., M.L.S., A.W., T.P., S.H., C.N., J.N., J.W., X.Z., J.M.R. and E.K. performed screenings, testing of biochemical activity, cell-based studies and animal studies; H.J.C., F.G., G.L., Ł.J. and M.J. performed the structural biology studies; W.Y., Y.Y. and Q.Z. synthesized compounds; M.L.G. and R.J.H.R. provided reagents and advised the study. All authors contributed to data analysis and writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
W.Y., Y.Y., F.G., Q.Z., J.G. and T.C. are co-inventors on a patent application for PRC1 inhibitors.
Additional information
Peer review information Nature Chemical Biology thanks Joshua Plotnik, Chi Wai Eric So and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Identification and characterization of fragment hit RB-1.
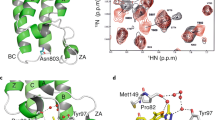
a, 1H-15N HSQC spectrum of 80 μM 15N RING1B-BMI1f (blue) superimposed onto 80 μM 15N RING1B-BMI1f with 2 mM RB-1 (red). Several residues with the largest chemical shift perturbations are labeled. b, In vitro ubiquitination assay showing inhibition of RING1B-BMI1f E3 ligase activity with RB-1. Purified HeLa nucleosomes were incubated with E1 (UBE1), E2 (UBCH5C), E3 (RING1B-BMI1f), ATP and Flag-tagged ubiquitin. Blots were probed with antibody against Flag. RING1B (K85E) corresponds to the assay with RING1B(K85E)-BMI1 mutant. Assay was repeated 2 times (top and bottom immunoblots). c, Mapping of chemical shift perturbations ΔHN (ppm) determined upon binding of 2 mM RB-1 to 80 μM RING1B-BMI1f. Residues are colored as follows: ΔHN < 0.01 (yellow); 0.01 ≤ ΔΗΝ < 0.04 (orange); ΔΗΝ ≥ 0.04 (red). BMI1 residues are in black. Location of K85 is labeled.
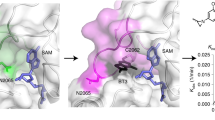
Extended Data Fig. 2 Structural characterization of the RING1B-BMI1f interaction with RB-2.
a, Superposition of the crystal structures of RING1B-BMI1f (colored gray) and RING1B-BMI1 complex (PDB 2CKL; RING1B is colored in magenta and BMI1 is blue). Positions of N- and C-termini are shown. b, Crystal structure determined for RING1B-BMI1f cocrystalized with RB-2. Electron density is contoured at 1 σ (blue) and selected residues are labeled. c, Crystal structure of RING1B-BMI1f cocrystalized with RB-2 shown in surface representation. Opened site exposing hydrophobic side chains of L80 and L100 (both in pale green) is shown. Surrounding positively charged residues are colored in pale blue. d, Analysis of chemical shift perturbations determined for 120 μM DCN RING1B-BMI1f upon binding with 500 μM RB-2. ΔHN has been calculated as \(\sqrt {({\Delta}\delta _{HN}^2 + 0.1 \ast {\Delta}\delta _N^2)}\) in ppm (top) and ΔCO is a difference in CO chemical shifts in ppm (bottom). e, Strips from 3D 1H-13C HSQC-NOESY spectra for 290 μM ILV 2H,13C,15N RING1B-BMI1f (black) and 120 μM ILV 2H,13C,15N RING1B-BMI1f with 500 μM RB-2 (blue). Assignment indicates NOEs between RING1B-BMI1f protein and RB-2). f, Labeling of the RB-2 protons. g, 20 lowest energy conformers of RING1B-BMI1f with bound RB-2. RING1B residues are in pale green and BMI1 are in gray. RB-2 is shown with magenta carbons. h, Binding of RB-2 to the wild-type RING1B-BMI1 and RING1B(L94A)-BMI1f point mutant. 1H-15N HSQC spectra for 60 μM 15N RING1B-BMI1f or 60 μM 15N RING1B(L94A)-BMI1f (shown in red) are titrated with 100 μM RB-2 (shown in blue).
Extended Data Fig. 3 Validation of the binding of RB-3 to RING1B-BMI1f and profiling the selectivity of RB-3.
a, Assigned 1H-13C HSQC spectrum of 60 μM 13C,15N RING1B-BMI1f (red) showing methyl group region superimposed onto the spectrum of 60 μM 13C,15N RING1B-BMI1f with 60 μM RB-3 (blue). b, Comparison of the binding of RB-3 and RB-nc using NMR. 1H-15N HSQC spectra of 60 μM 15N RING1B-BMI1f (red) titrated with 100 μM RB-nc (green), 400 μM RB-nc (blue) and 100 μM RB-3 (black). Of note, 100 μM RB-3 results in almost complete saturation of chemical shift perturbations with 60 μM 15N RING1B-BMI1f. c, In vitro ubiquitination assay showing effect of RB-nc on RING1B-BMI1 E3 ligase activity. H2Aub was detected using Western blot and H3 was used as loading control. Lane with no E3 depicts sample without RING1B-BMI1f serving as negative control. Assay was repeated two times. d, 1H-15N HSQC spectrum of 60 μM 15N RING1B-BMI1f with 5% DMSO (black) superimposed onto 60 μM 15N RING1B-BMI1f with 60 μM (blue) and 120 μM RB-3 (red). e, 1H-15N HSQC spectrum of 60 μM 15N RING1A-BMI1f with 5% DMSO (black) superimposed onto 60 μM 15N RING1A-BMI1f with 60 μM (blue) and 120 μM RB-3 (red). f, In vitro ubiquitination assay showing the effect of RB-3 on RING1A-BMI1f and RING1B-BMI1f activity. H2Aub was detected using Western blot and H3 was used as loading control. Assays were repeated two times. g, In vitro ubiquitination assays showing no activity of RB-3 on BRCA1-BARD1, TRIM37 and RNF168. H2Aub is detected using Western blot and H3 or H2A was used as loading controls. Lanes with no E3 depicts samples without BRCA1-BARD1, TRIM37 or RNF168, respectively, serving as negative controls. Assays were repeated two times.
Extended Data Fig. 4 Development and characterization of biotinylated RING1B probe compounds.
a, structures of RB-3-biot and RB-nc-biot. b, 1H-15N HSQC spectra of 60 μM 15N RING1B-BMI1f (green) titrated with 60 μM RB-3-biot or RB-nc-biot (red) and 120 μM RB-3-biot or RB-nc-biot (blue). c, Pull-down from HEK293T cells with RB-3-biot or RB-nc-biot followed by detection of RING1B or BMI1 using Western Blot analysis. Two controls were used in the pull-down assay, DMSO or biotinylated histone H3 peptide (biotin-H3), to assess non-specific binding. Assays were repeated two times.
Extended Data Fig. 5 RB-3 inhibits H2Aub in cancer cell lines.
a, Evaluation of BMI1 and RING1B knockdown, and treatment with RB-3 in K562 cells. Left western blot analysis in K562 cells transfected with BMI1/RING1B siRNAs for 4 d. Right immunoblots of K562 cells treated with indicated doses of RB-3 for 4 d. Representative blots of two independent experiments are shown. b, Evaluation of BMI1 and RING1B knockdown, and treatment with RB-3 in HeLa cells. Left Western blot in HeLa cells transfected with BMI1/RING1B siRNAs for 4 d. Right western blot analysis in HeLa cells treated with indicated doses of RB-3 for 4 d. Representative blots of two independent experiments are shown. c, Activity of RB-3 and RB-nc in MOLM13 cells treated for 4 d with indicated doses of compounds. Representative blots of two independent experiments are shown. d, Activity of RB-3 in MV4;11 cells treated for 4 d. Representative blots of two independent experiments are shown.
Extended Data Fig. 6 Long-term effect of treatment of TEX cells with RB-3.
a, Western blot detection of H2Aub in TEX cells treated with increasing doses of RB-3 and RB-nc for 21 days. Representative blot out of two replicates. b, Western blot detection of total cellular ubiquitination levels using ubiquitin specific antibody in TEX cells treated with RB-3 for 21 days. Representative blot out of three replicates. c, Flow cytometry analysis of CD34 in TEX cells treated with RB-3 for 7 days. On day 7, cells were stained with APC/CY7-conjugated human anti-CD34 antibody and analyzed by FACS. Representative histograms of two independent experiments. d, Flow cytometry analysis of CD34 and CD38 in TEX cells treated with RB-3 (top panel) and RB-nc (bottom panel) for 21 days. On day 21, cells were stained with APC/CY7-conjugated human CD34 and PE-conjugated CD38 antibodies and analyzed by FACS. Representative histograms of two independent experiments. e,f, Flow cytometry analysis of myeloid differentiation marker CD11b/ITGAM in TEX cells treated with RB-3 for 7 days (e) and with RB-3 (f, top panel) and RB-nc (f, bottom panel) for 21 days. Cells were stained with Pacific Blue human CD11b antibody and analyzed by FACS. Representative histograms of two independent experiments. g, Flow cytometry analysis of dendritic cells differentiation marker CD86/B7-2 in TEX cells treated with RB-3 (top panel) and RB-nc (bottom panel) for 21 days. On day 21, cells were stained with Super Bight 436 conjugated human CD86 antibody and analyzed by FACS. Representative histograms of two independent experiments.
Extended Data Fig. 7 RB-3 regulates expression of target genes and impairs enrichment of H2Aub and RING1B on target gene promoter regions in TEX cells.
a, qRT–PCR showing time dependent changes in transcript levels of CD34, C/EBPα, CD-86 upon treatment with 25 μM RB-3 and RB-nc. Representative data out of two replicates. b, Analysis of the H2Aub and H3 levels, and binding of RING1B to the two promoter regions of C/EBPα in TEX cells treated with RB-nc and DMSO for 8 days using ChIP assay. Representative data out of two replicates. c, Analysis of the H2Aub and H3 levels, and binding of RING1B to the two promoter regions of CD34 in TEX cells treated with RB-3 (red), RB-nc (blue) and DMSO (black) for 8 days using ChIP assay. Representative data out of two replicates. d, Analysis of the H2Aub and H3 levels, and binding of RING1B to the two promoter regions of ITGAM in TEX cells treated with RB-3 (red), RB-nc (blue) and DMSO (black) for 8 days using ChIP assay. Representative data out of two replicates. Two promoter region primers between 1 to 2 kb upstream of the transcription start site (TSS) for each of CD34, C/EBPα and ITGAM promoter were selected and analyzed (panels b, c, d). e, Average genome-wide occupancy of H2Aub around the transcription start sites (TSS) and genomic regions in TEX cells determined from ChIP-seq experiment. TEX cells were treated with DMSO and 25 μM RB-3 for 6 days.
Extended Data Fig. 8 Comparison of the activity of PTC209 and RB-3 in TEX cells.
a, Graph showing effect of PTC209 on TEX cells proliferation. TEX cells were treated with the indicated concentrations of PTC209 for 4 days and processed for cell proliferation analysis. Experiment was performed 2 times, GI50 is mean ± s.d. b, western blot showing levels of H2Aub, H2Bub, H2A, H3 and BMI1 in TEX cells treated for 4 days with indicated doses of RB-3 (left) and PTC209 (right). Representative blots from two independent experiments. c, qRT–PCR indicating levels of C/EBPα upon 4 d treatment with indicated doses of RB-3 and PTC209. Representative data from two replicates. d, qRT–PCR indicating levels of CD34 upon 4 d treatment with indicated doses of RB-3 and PTC209. Representative data from two replicates.
Extended Data Fig. 9 RB-3 has no effect on normal CD34+ cells.
a, Quantitation of colony numbers (CFU) upon treatment of cord blood CD34+ cells from healthy donors with RB-3. Experiments were performed two times and representative data are mean ± s.d. and analyzed by two-tailed t-test; ns – not significant. b, Population of CD34+ cells upon treatment with RB-3. Experiments were performed two times and representative data are mean ± s.d. and analyzed by two-tailed t-test; ns – not significant. c, Flow cytometry analysis of CD34 and CD38. Representative histograms of two independent experiments. d, Effect of RB-3 on various populations of mature cells with representative colony pictures. CFU-GM, granulocyte–macrophage progenitors; CFU-GEMM, oligopotential progenitors; BFU-E, burst-forming unit-erythroid cells; CFU-E, colony-forming unit-erythroid cells. Experiments were performed two times and representative data are mean ± s.d. and analyzed by two-tailed t-test; ns – not significant. e, Cell morphology of CD34+ cells treated with RB-3 analyzed by Wright Giemsa staining. Representative slides are shown from two independent experiments.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Figs. 1–7 and Note.
Source data
Source Data Fig. 1
Uncropped gels.
Source Data Fig. 2
Uncropped gels.
Source Data Fig. 3
Uncropped gels.
Source Data Fig. 4
Uncropped gels.
Source Data Fig. 5
Uncropped gels.
Source Data Fig. 6
Uncropped gels.
Source Data Extended Data Fig. 1
Uncropped gels.
Source Data Extended Data Fig. 3
Uncropped gels.
Source Data Extended Data Fig. 4
Uncropped gels.
Source Data Extended Data Fig. 5
Uncropped gels.
Source Data Extended Data Fig. 6
Uncropped gels.
Source Data Extended Data Fig. 8
Uncropped gels.
Rights and permissions
About this article
Cite this article
Shukla, S., Ying, W., Gray, F. et al. Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain. Nat Chem Biol 17, 784–793 (2021). https://doi.org/10.1038/s41589-021-00815-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00815-5
This article is cited by
-
The cross talk of ubiquitination and chemotherapy tolerance in colorectal cancer
Journal of Cancer Research and Clinical Oncology (2024)
-
Changes in PRC1 activity during interphase modulate lineage transition in pluripotent cells
Nature Communications (2023)
-
SUZ12 inhibition attenuates cell proliferation of glioblastoma via post-translational regulation of CDKN1B
Genes & Genomics (2023)
-
Fighting PRC1 via the RING
Nature Chemical Biology (2021)