Abstract
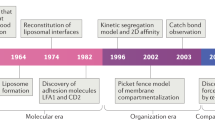
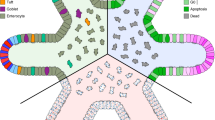
Multicellular organisms depend on physical cell–cell interactions to control physiological processes such as tissue formation, neurotransmission and immune response. These intercellular binding events can be both highly dynamic in their duration and complex in their composition, involving the participation of many different surface and intracellular biomolecules. Untangling the intricacy of these interactions and the signaling pathways they modulate has greatly improved insight into the biological processes that ensue upon cell–cell engagement and has led to the development of protein- and cell-based therapeutics. The importance of monitoring physical cell–cell interactions has inspired the development of several emerging approaches that effectively interrogate cell–cell interfaces with molecular-level detail. Specifically, the merging of chemistry- and biology-based technologies to deconstruct the complexity of cell–cell interactions has provided new avenues for understanding cell–cell interaction biology and opened opportunities for therapeutic development.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Yamada, S. & Nelson, W. J. Synapses: sites of cell recognition, adhesion and functional specification. Annu. Rev. Biochem. 76, 267–294 (2007).
Belardi, B., Son, S., Felce, J. H., Dustin, M. L. & Fletcher, D. A. Cell–cell interfaces as specialized compartments directing cell function. Nat. Rev. Mol. Cell Biol. 21, 750–764 (2020).
Darvin, P., Toor, S. M., Sasidharan Nair, V. & Elkord, E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp. Mol. Med. 50, 1–11 (2018).
Rafiq, S., Hackett, C. S. & Brentjens, R. J. Engineering strategies to overcome the current roadblocks in CAR T-cell therapy. Nat. Rev. Clin. Oncol. 17, 147–167 (2020).
Huse, M. Mechanical forces in the immune system. Nat. Rev. Immunol. 17, 679–690 (2017).
Polacheck, W. J. & Chen, C. S. Measuring cell-generated forces: a guide to the available tools. Nat. Methods 13, 415–423 (2016).
Armingol, E., Officer, A., Harismendy, O. & Lewis, N. E. Deciphering cell-cell interactions and communication from gene expression. Nat. Rev. Genet. 22, 71–88 (2021).
Wilson, H. V. On some phenomena of coalescence and regeneration in sponges. J. Exp. Zool. 5, 245–258 (1907).
Townes, P. L. & Holtfreter, J. Directed movements and selective adhesion of embryonic amphibian cells. J. Exp. Zool. 128, 53–120 (1955).
Moscona, A. & Moscona, H. The dissociation and aggregation of cells from organ rudiments of the early chick embryo. J. Anat. 86, 287–301 (1952).
Davidson, M. W. & Abramowitz, M. in Encyclopedia of Imaging Science and Technology (ed. Hornak, J.) 1106–1141 (Wiley, 2002).
Wollman, A. J. M., Nudd, R., Hedlund, E. G. & Leake, M. C. From Animaculum to single molecules: 300 years of the light microscope. Open Biol. 5, 150019 (2015).
Werner, M., von Wasielewski, R. & Komminoth, P. Antigen retrieval, signal amplification and intensification in immunohistochemistry. Histochem. Cell Biol. 105, 253–260 (1996).
Stack, E. C., Wang, C. C., Roman, K. A. & Hoyt, C. C. Multiplexed immunohistochemistry, imaging and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 70, 46–58 (2014).
Wang, L., Frei, M. S., Salim, A. & Johnsson, K. Small-molecule fluorescent probes for live-cell super-resolution microscopy. J. Am. Chem. Soc. 141, 2770–2781 (2019).
Specht, E. A., Braselmann, E. & Palmer, A. E. A critical and comparative review of fluorescent tools for live-cell imaging. Annu. Rev. Physiol. 79, 93–117 (2017).
Yap, A. S., Michael, M. & Parton, R. G. Seeing and believing: recent advances in imaging cell–cell interactions. F1000Res. 4, 273 (2015).
Baharlou, H., Canete, N. P., Cunningham, A. L., Harman, A. N. & Patrick, E. Mass cytometry imaging for the study of human diseases—applications and data analysis strategies. Front. Immunol. 10, 2657 (2019).
Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020).
Barteneva, N. S., Fasler-Kan, E. & Vorobjev, I. A. Imaging flow cytometry: coping with heterogeneity in biological systems. J. Histochem. Cytochem. 60, 723–733 (2012).
Burel, J. G. et al. Circulating T cell–monocyte complexes are markers of immune perturbations. eLife 8, e46045 (2019).
Popescu, D. M. et al. Decoding human fetal liver haematopoiesis. Nature 574, 365–371 (2019).
Groves, J. T. & Dustin, M. L. Supported planar bilayers in studies on immune cell adhesion and communication. J. Immunol. Methods 278, 19–32 (2003).
Monks, C. R., Freiberg, B. A., Kupfer, H., Sciaky, N. & Kupfer, A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 395, 82–86 (1998).
Grakoui, A. et al. The immunological synapse: a molecular machine controlling T cell activation. Science 285, 221–227 (1999).
Kaizuka, Y., Douglass, A. D., Varma, R., Dustin, M. L. & Vale, R. D. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc. Natl Acad. Sci. USA 104, 20296–20301 (2007).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Dani, A., Huang, B., Bergan, J., Dulac, C. & Zhuang, X. Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856 (2010).
Wu, Y., Kanchanawong, P. & Zaidel-Bar, R. Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions. Dev. Cell 32, 139–154 (2015).
Chamma, I. et al. Mapping the dynamics and nanoscale organization of synaptic adhesion proteins using monomeric streptavidin. Nat. Commun. 7, 10773 (2016).
Ries, J., Kaplan, C., Platonova, E., Eghlidi, H. & Ewers, H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584 (2012).
Rothbauer, U. et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889 (2006).
Beghein, E. & Gettemans, J. Nanobody technology: a versatile toolkit for microscopic imaging, protein–protein interaction analysis, and protein function exploration. Front. Immunol. 8, 771 (2017).
Feinberg, E. H. et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363 (2008).
Kim, J. et al. mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat. Methods 9, 96–102 (2011).
Macpherson, L. J. et al. Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat. Commun. 6, 10024 (2015).
Liu, D. S., Loh, K. H., Lam, S. S., White, K. A. & Ting, A. Y. Imaging trans-cellular neurexin–neuroligin interactions by enzymatic probe ligation. PLoS ONE 8, e52823 (2013).
Martell, J. D. et al. A split horseradish peroxidase for the detection of intercellular protein–protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 34, 774–780 (2016).
Carpenter, M. A. et al. Protein proximity observed using fluorogen activating protein and dye activated by proximal anchoring (FAP–DAPA) system. ACS Chem. Biol. 15, 2433–2443 (2020).
Stein, J. V. & Gonzalez, S. F. Dynamic intravital imaging of cell–cell interactions in the lymph node. J. Allergy Clin. Immunol. 139, 12–20 (2017).
Miller, M. J., Wei, S. H., Parker, I. & Cahalan, M. D. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science 296, 1869–1873 (2002).
Miller, M. J., Hejazi, A. S., Wei, S. H., Cahalan, M. D. & Parker, I. T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc. Natl Acad. Sci. USA 101, 998–1003 (2004).
Pasqual, G. et al. Monitoring T cell–dendritic cell interactions in vivo by intercellular enzymatic labelling. Nature 553, 496–500 (2018).
Ge, Y. et al. Enzyme-mediated intercellular proximity labeling for detecting cell–cell interactions. J. Am. Chem. Soc. 141, 1833–1837 (2019).
Liu, Q. et al. A proximity-tagging system to identify membrane protein–protein interactions. Nat. Methods 15, 715–722 (2018).
Liu, Z. L. et al. Detecting tumor antigen-specific T cells via interaction-dependent fucosyl-biotinylation. Cell 183, 1117–1133 (2020).
Piersimoni, L. & Sinz, A. Cross-linking/mass spectrometry at the crossroads. Anal. Bioanal. Chem. 412, 5981–5987 (2020).
Gonzalez-Lozano, M. A. et al. Stitching the synapse: cross-linking mass spectrometry into resolving synaptic protein interactions. Sci. Adv. 6, eaax5783 (2020).
Cho, K. F. et al. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat. Protoc. 15, 3971–3999 (2020).
Qin, W., Cho, K. F., Cavanagh, P. E. & Ting, A. Y. Deciphering molecular interactions by proximity labeling. Nat. Methods 18, 133–143 (2021).
Kim, D. I. et al. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl Acad. Sci. USA 111, E2453–E2461 (2014).
Martell, J. D. et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30, 1143–1148 (2012).
Rhee, H. W. et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013).
Loh, K. H. et al. Proteomic analysis of unbounded cellular compartments: synaptic clefts. Cell 166, 1295–1307 (2016).
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
Shafraz, O., Xie, B., Yamada, S. & Sivasankar, S. Mapping transmembrane binding partners for E-cadherin ectodomains. Proc. Natl Acad. Sci. USA 117, 31157–31165 (2020).
Kwak, C. et al. Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proc. Natl Acad. Sci. USA 117, 12109–12120 (2020).
Cho, K. F. et al. Split-TurboID enables contact-dependent proximity labeling in cells. Proc. Natl Acad. Sci. USA 117, 12143–12154 (2020).
Samavarchi-Tehrani, P., Samson, R. & Gingras, A. C. Proximity dependent biotinylation: key enzymes and adaptation to proteomics approaches. Mol. Cell Proteom. 19, 757–773 (2020).
Prier, C. K., Rankic, D. A. & MacMillan, D. W. C. Visible light photoredox catalysis with transition metal complexes: applications in organic synthesis. Chem. Rev. 113, 5322–5363 (2013).
Geri, J. B. et al. Microenvironment mapping via Dexter energy transfer on immune cells. Science 367, 1091–1097 (2020).
Patel, S. J. et al. Identification of essential genes for cancer immunotherapy. Nature 548, 537–542 (2017).
Vredevoogd, D. W. et al. Augmenting immunotherapy impact by lowering tumor TNF cytotoxicity threshold. Cell 178, 585–599 (2019).
Kula, T. et al. T-Scan: a genome-wide method for the systematic discovery of T cell epitopes. Cell 178, 1016–1028 (2019).
Morsut, L. et al. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell 164, 780–791 (2016).
Toda, S., Blauch, L. R., Tang, S. K. Y., Morsut, L. & Lim, W. A. Programming self-organizing multicellular structures with synthetic cell–cell signaling. Science 361, 156–162 (2018).
Roybal, K. T. et al. Engineering T cells with customized therapeutic response programs using synthetic notch receptors. Cell 167, 419–432 (2016).
Tang, R. et al. A versatile system to record cell–cell interactions. eLife 9, e61080 (2020).
Talay, M. et al. Transsynaptic mapping of second-order taste neurons in flies by trans-Tango. Neuron 96, 783–795 (2017).
Waldman, A. D., Fritz, J. M. & Lenardo, M. J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668 (2020).
Kontermann, R. E. & Brinkmann, U. Bispecific antibodies. Drug Discov. Today 20, 838–847 (2015).
Labrijn, A. F., Janmaat, M. L., Reichert, J. M. & Parren, P. Bispecific antibodies: a mechanistic review of the pipeline. Nat. Rev. Drug Discov. 18, 585–608 (2019).
Labanieh, L., Majzner, R. G. & Mackall, C. L. Programming CAR-T cells to kill cancer. Nat. Biomed. Eng. 2, 377–391 (2018).
Bommareddy, P. K., Shettigar, M. & Kaufman, H. L. Integrating oncolytic viruses in combination cancer immunotherapy. Nat. Rev. Immunol. 18, 498–513 (2018).
Twumasi-Boateng, K., Pettigrew, J. L., Kwok, Y. Y. E., Bell, J. C. & Nelson, B. H. Oncolytic viruses as engineering platforms for combination immunotherapy. Nat. Rev. Cancer 18, 419–432 (2018).
Dura, B. et al. Longitudinal multiparameter assay of lymphocyte interactions from onset by microfluidic cell pairing and culture. Proc. Natl Acad. Sci. USA 113, E3599–E3608 (2016).
Ben-Moshe, S. et al. Spatial sorting enables comprehensive characterization of liver zonation. Nat. Metab. 1, 899–911 (2019).
Moor, A. E. et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell 175, 1156–1167 (2018).
Giladi, A. et al. Dissecting cellular crosstalk by sequencing physically interacting cells. Nat. Biotechnol. 38, 629–637 (2020).
Williams, J. Z. et al. Precise T cell recognition programs designed by transcriptionally linking multiple receptors. Science 370, 1099–1104 (2020).
Lajoie, M. J. et al. Designed protein logic to target cells with precise combinations of surface antigens. Science 369, 1637–1643 (2020).
Acknowledgements
We thank Y. Zheng of Yizheng Illustrations for figure design work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
T.J.B., T.R.R., O.O.F. and R.C.O. are employees of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA.
Additional information
Peer review information Nature Chemical Biology thanks Cheng Zhu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bechtel, T.J., Reyes-Robles, T., Fadeyi, O.O. et al. Strategies for monitoring cell–cell interactions. Nat Chem Biol 17, 641–652 (2021). https://doi.org/10.1038/s41589-021-00790-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00790-x
This article is cited by
-
Universal recording of immune cell interactions in vivo
Nature (2024)
-
The diversification of methods for studying cell–cell interactions and communication
Nature Reviews Genetics (2024)
-
Proximity extracellular protein-protein interaction analysis of EGFR using AirID-conjugated fragment of antigen binding
Nature Communications (2023)
-
Targeted activation in localized protein environments via deep red photoredox catalysis
Nature Chemistry (2023)
-
Secretory GFP reconstitution labeling of neighboring cells interrogates cell–cell interactions in metastatic niches
Nature Communications (2023)