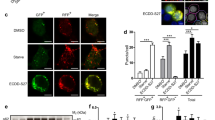
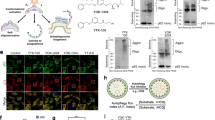
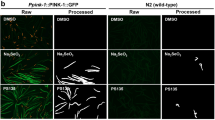
Abstract
Autophagy is implicated in a wide range of (patho)physiological processes including maintenance of cellular homeostasis, neurodegenerative disorders, aging and cancer. As such, small molecule autophagy modulators are in great demand, both for their ability to act as tools to better understand this essential process and as potential therapeutics. Despite substantial advances in the field, major challenges remain in the development and comprehensive characterization of probes that are specific to autophagy. In this Review, we discuss recent developments in autophagy-modulating small molecules, including the specific challenges faced in the development of activators and inhibitors, and recommend guidelines for their use. Finally, we discuss the potential to hijack the process for targeted protein degradation, an area of great importance in chemical biology and drug discovery.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wong, E. et al. Molecular determinants of selective clearance of protein inclusions by autophagy. Nat. Commun. 3, 1240 (2012).
Fujioka, Y. et al. Phase separation organizes the site of autophagosome formation. Nature 578, 301–305 (2020).
Dikic, I. & Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018).
Pohl, C. & Dikic, I. Cellular quality control by the ubiquitin–proteasome system and autophagy. Science 366, 818–822 (2019).
Li, W. W., Li, J. & Bao, J. K. Microautophagy: lesser-known self-eating. Cell. Mol. Life Sci. 69, 1125–1136 (2012).
Kaushik, S. & Cuervo, A. M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018).
Levine, B. & Kroemer, G. Biological functions of autophagy genes: a disease perspective. Cell 176, 11–42 (2019).
Galluzzi, L., Bravo-San Pedro, J. M., Levine, B., Green, D. R. & Kroemer, G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 16, 487–511 (2017).
Hosokawa, N. et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 (2009).
Jung, C. H. et al. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009).
Kaufmann, A., Beier, V., Franquelim, H. G. & Wollert, T. Molecular mechanism of autophagic membrane–scaffold assembly and disassembly. Cell 156, 469–481 (2014).
Itakura, E., Kishi-Itakura, C. & Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012).
Galluzzi, L. & Green, D. R. Autophagy-independent functions of the autophagy machinery. Cell 177, 1682–1699 (2019).
Levy, J. M. M., Towers, C. G. & Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542 (2017).
Nixon, R. A. The role of autophagy in neurodegenerative disease. Nat. Med. 19, 983–997 (2013).
Sharma, V. et al. Selective autophagy and xenophagy in infection and disease. Front. Cell Dev. Biol. 6, 147 (2018).
Bonam, S. R., Wang, F. & Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 18, 923–948 (2019).
Fernández, Á. F. et al. Disruption of the beclin 1–BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140 (2018).
Perera, R. M. et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nature 524, 361–382 (2015).
Yamamoto, K. et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581, 100–105 (2020). This paper describes a new role for autophagy in sustaining pancreatic cancer by degrading MHC-I and promoting immune evasion.
Cassidy, L. D. et al. Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nat. Commun. 11, 307 (2020).
Pietrocola, F. et al. Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell 30, 147–160 (2016).
Workman, P. & Collins, I. Probing the probes: fitness factors for small molecule tools. Chem. Biol. 17, 561–577 (2010).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016).
Dong, Y. et al. Autophagy modulator scoring system: a user-friendly tool for quantitative analysis of methodological integrity of chemical autophagy modulator studies. Autophagy 16, 195–202 (2020).
Mizushima, N. & Murphy, L. O. Autophagy assays for biological discovery and therapeutic development. Trends Biochem. Sci. 45, 1080–1093 (2020).
Kaizuka, T. et al. An autophagic flux probe that releases an internal control. Mol. Cell 64, 835–849 (2016). This paper describes a probe that can reliably be used to identify modulators of autophagic flux.
Nanduri, R. et al. AutophagySMDB: a curated database of small molecules that modulate protein targets regulating autophagy. Autophagy 15, 1280–1295 (2019).
Gwinn, D. M. et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 (2008).
Sabatini, D. M., Erdjument-Bromage, H., Lui, M., Tempst, P. & Snyder, S. H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78, 35–43 (1994).
Blommaart, E. F. C., Luiken, J. J. F. P., Blommaart, P. J. E., Van Woerkom, G. M. & Meijer, A. J. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270, 2320–2326 (1995).
Liu, Q. et al. Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benzo[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. J. Med. Chem. 53, 7146–7155 (2010).
Chresta, C. M. et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 70, 288–298 (2010).
Wang, Y. et al. Metformin induces autophagy and G0/G1 phase cell cycle arrest in myeloma by targeting the AMPK/mTORC1 and mTORC2 pathways. J. Exp. Clin. Cancer Res. 37, 63 (2018).
Sarkar, S., Davies, J. E., Huang, Z., Tunnacliffe, A. & Rubinsztein, D. C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and α-synuclein. J. Biol. Chem. 282, 5641–5652 (2007).
Sarkar, S. et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat. Chem. Biol. 3, 331–338 (2007).
Kuo, S.-Y. et al. Small-molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics. Proc. Natl Acad. Sci. USA 112, E4281–E4287 (2015).
DeBosch, B. J. et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci. Signal. 9, ra21 (2016).
Rusmini, P. et al. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy 15, 631–651 (2019).
Sarkar, S. et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 170, 1101–1111 (2005).
Siddiqi, F. H. et al. Felodipine induces autophagy in mouse brains with pharmacokinetics amenable to repurposing. Nat. Commun. 10, 1817 (2019).
Chung, C. Y. S. et al. Covalent targeting of the vacuolar H+-ATPase activates autophagy via mTORC1 inhibition. Nat. Chem. Biol. 15, 776–785 (2019). This paper describes a covalent modifier of the v-ATPase that induces autophagy.
Lim, C. Y. et al. ER–lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann–Pick type C. Nat. Cell Biol. 21, 1206–1218 (2019).
Burgett, A. W. G. et al. Natural products reveal cancer cell dependence on oxysterol-binding proteins. Nat. Chem. Biol. 7, 639–647 (2011).
Rosato, A. S. et al. TRPML1 links lysosomal calcium to autophagosome biogenesis through the activation of the CaMKK1B/VPS34 pathway. Nat. Commun. 10, 5630 (2019).
Medina, D. L. et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288–299 (2015).
Chen, C. et al. A small molecule restores function to TRPML1 mutant isoforms responsible for mucolipidosis type IV. Nat. Commun. 5, 4681 (2014).
Shen, D. et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 3, 731 (2012).
Laraia, L., McKenzie, G., Spring, D. R., Venkitaraman, A. R. & Huggins, D. J. Overcoming chemical, biological, and computational challenges in the development of inhibitors targeting protein–protein interactions. Chem. Biol. 22, 689–703 (2015).
Chiang, W. C. et al. High-throughput screens to identify autophagy inducers that function by disrupting beclin 1/Bcl-2 binding. ACS Chem. Biol. 13, 2247–2260 (2018).
Shoji-Kawata, S. et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206 (2013). This paper describes the identification of a peptide inhibitor of a protein–protein interaction that leads to autophagy induction.
Georgakopoulos, N. D., Wells, G. & Campanella, M. The pharmacological regulation of cellular mitophagy. Nat. Chem. Biol. 13, 136–146 (2017).
Robke, L. et al. Discovery of the novel autophagy inhibitor aumitin that targets mitochondrial complex I. Chem. Sci. 9, 3014–3022 (2018).
Chu, C. T. et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 15, 1197–1205 (2013).
Bosc, C. et al. Autophagy regulates fatty acid availability for oxidative phosphorylation through mitochondria–endoplasmic reticulum contact sites. Nat. Commun. 11, 4056 (2020).
Rusilowicz-Jones, E. V. et al. USP30 sets a trigger threshold for PINK1–PARKIN amplification of mitochondrial ubiquitylation. Life Sci. Alliance 3, e202000768 (2020). This paper describes one of the first examples of a mitophagy-enhancing compound that does not disrupt the mitochondrial membrane potential.
Heckmann, B. L., Yang, X., Zhang, X. & Liu, J. The autophagic inhibitor 3-methyladenine potently stimulates PKA-dependent lipolysis in adipocytes. Br. J. Pharmacol. 168, 163–171 (2013).
Sarkaria, J. N. et al. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 58, 4375–4382 (1998).
Robke, L. et al. Phenotypic identification of a novel autophagy inhibitor chemotype targeting lipid kinase VPS34. Angew. Chem. Int. Ed. 56, 8153–8157 (2017).
Foley, D. J. et al. Phenotyping reveals the targets of a pseudo-natural product autophagy inhibitor. Angew. Chem. Int. Ed. 59, 12470–12476 (2020).
Ronan, B. et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10, 1013–1019 (2014). This paper describes the discovery of the most potent and selective VPS34 inhibitor to date.
Bago, R. et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 463, 413–427 (2014).
Noman, M. Z. et al. Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti–PD-1/PD-L1 immunotherapy. Sci. Adv. 6, eaax7881 (2020).
Pavlinov, I., Salkovski, M. & Aldrich, L. N. Beclin 1–ATG14L protein–protein interaction inhibitor selectively inhibits autophagy through disruption of VPS34 complex I. J. Am. Chem. Soc. 142, 8174–8182 (2020).
Petherick, K. J. et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 290, 11376–11383 (2015).
Egan, D. F. et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 59, 285–297 (2015).
Martin, K. R. et al. A potent and selective ULK1 inhibitor suppresses autophagy and sensitizes cancer cells to nutrient stress. iScience 8, 74–84 (2018). This paper describes the identification of the most potent and selective ULK1 inhibitor to date.
Bosc, D. et al. A new quinoline-based chemical probe inhibits the autophagy-related cysteine protease ATG4B. Sci. Rep. 8, 11653 (2018).
Qiu, Z. et al. Discovery of fluoromethylketone-based peptidomimetics as covalent ATG4B (autophagin-1) inhibitors. ACS Med. Chem. Lett. 7, 802–806 (2016).
Huang, S. C. et al. Discovery and optimization of pyrazolopyrimidine sulfamates as ATG7 inhibitors. Bioorg. Med. Chem. 28, 115681 (2020).
Laraia, L. et al. The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis. Nat. Chem. Biol. 15, 710–720 (2019). This paper describes a role for cholesterol transfer proteins in autophagosome biogenesis and provides tool compounds for inhibiting them.
Wijdeven, R. H. et al. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 7, 11808 (2016).
Samie, M. et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev. Cell 26, 511–524 (2013).
Njomen, E. & Tepe, J. J. Regulation of autophagic flux by the 20S proteasome. Cell Chem. Biol. 26, 1283–1294 (2019).
Bowman, E. J., Sieberst, A. & Altendorft, K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl Acad. Sci. USA 85, 7972–7976 (1988).
Huss, M. et al. Concanamycin A, the specific inhibitor of V-ATPases, binds to the Vo subunit c. J. Biol. Chem. 277, 40544–40548 (2002).
Xie, X. S. et al. Salicylihalamide A inhibits the Vo sector of the V-ATPase through a mechanism distinct from bafilomycin A1. J. Biol. Chem. 279, 19755–19763 (2004).
Boyd, M. R. et al. Discovery of a novel antitumor benzolactone enamide class that selectively inhibits mammalian vacuolar-type H+-ATPases. J. Pharmacol. Exp. Ther. 297, 114–120 (2001).
Sørensen, M. G., Henriksen, K., Neutzsky-Wulff, A. V., Dziegiel, M. H. & Karsdal, M. A. Diphyllin, a novel and naturally potent v-ATPase inhibitor, abrogates acidification of the osteoclastic resorption lacunae and bone resorption. J. Bone Miner. Res. 22, 1640–1648 (2007).
Wang, Y. et al. Pharmacological targeting of vacuolar H+-ATPase via subunit V1G combats multidrug-resistant cancer. Cell Chem. Biol. 27, 1359–1370 (2020).
Aldrich, L. N. et al. Discovery of a small-molecule probe for v-ATPase function. J. Am. Chem. Soc. 137, 5563–5568 (2015).
Kapishnikov, S. et al. Mode of action of quinoline antimalarial drugs in red blood cells infected by Plasmodium falciparum revealed in vivo. Proc. Natl Acad. Sci. USA 116, 22946–22952 (2019).
Sumpter, M. D., Tatro, L. S., Stoecker, W. V. & Rader, R. K. Evidence for risk of cardiomyopathy with hydroxychloroquine. Lupus 21, 1594–1596 (2012).
Mauthe, M. et al. Chloroquine inhibits autophagic flux by decreasing autophagosome–lysosome fusion. Autophagy 14, 1435–1455 (2018).
Ashoor, R., Yafawi, R., Jessen, B. & Lu, S. The contribution of lysosomotropism to autophagy perturbation. PLoS ONE 8, e82481 (2013).
Kornhuber, J. et al. Identification of new functional inhibitors of acid sphingomyelinase using a structure–property–activity relation model. J. Med. Chem. 51, 219–237 (2008).
Laraia, L. et al. Image-based morphological profiling identifies a lysosomotropic, iron-sequestering autophagy inhibitor. Angew. Chem. Int. Ed. 59, 5721–5729 (2020).
Laraia, L. et al. Discovery of novel cinchona-alkaloid-inspired oxazatwistane autophagy inhibitors. Angew. Chem. Int. Ed. 56, 2177–2182 (2017).
Kimura, S., Noda, T. & Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007).
Burslem, G. M. & Crews, C. M. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 181, 102–114 (2020).
Ito, C. et al. Endogenous nitrated nucleotide is a key mediator of autophagy and innate defense against bacteria. Mol. Cell 52, 794–804 (2013).
Takahashi, D. et al. AUTACs: cargo-specific degraders using selective autophagy. Mol. Cell 76, 797–810 (2019). This paper describes the first example of targeting proteins to the autophagosome for degradation using small molecules.
Fan, X., Jin, W. Y., Lu, J., Wang, J. & Wang, Y. T. Rapid and reversible knockdown of endogenous proteins by peptide-directed lysosomal degradation. Nat. Neurosci. 17, 471–480 (2014).
Bauer, P. O. et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat. Biotechnol. 28, 256–263 (2010).
Zhou, Y.-F. et al. The peptide-directed lysosomal degradation of CDK5 exerts therapeutic effects against stroke. Aging Dis. 10, 1140–1145 (2019).
Wang, H. et al. HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity. Nat. Chem. Biol. 15, 42–50 (2019).
Gary-Bobo, M., Nirdé, P., Jeanjean, A., Morère, A. & Garcia, M. Mannose 6-phosphate receptor targeting and its applications in human diseases. Curr. Med. Chem. 14, 2945–2953 (2007).
Zhu, Y. et al. Conjugation of mannose 6-phosphate-containing oligosaccharides to acid α-glucosidase improves the clearance of glycogen in Pompe mice. J. Biol. Chem. 279, 50336–50341 (2004).
Banik, S. M. et al. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 584, 291–297 (2020). This paper describes the first example of targeting extracellular proteins to the lysosome for degradation using designed molecules.
Anguiano, J. et al. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat. Chem. Biol. 9, 374–382 (2013).
Acknowledgements
We gratefully acknowledge the Novo Nordisk Foundation, the Independent Research Fund Denmark, the Carlsberg Foundation and DTU for financial support to L.L. T.W.-E. acknowledges DTU for a PhD fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemical Biology thanks Sovan Sarkar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Whitmarsh-Everiss, T., Laraia, L. Small molecule probes for targeting autophagy. Nat Chem Biol 17, 653–664 (2021). https://doi.org/10.1038/s41589-021-00768-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00768-9
This article is cited by
-
S670, an amide derivative of 3-O-acetyl-11-keto-β-boswellic acid, induces ferroptosis in human glioblastoma cells by generating ROS and inhibiting STX17-mediated fusion of autophagosome and lysosome
Acta Pharmacologica Sinica (2024)
-
Endothelial autophagy blockade fosters anti-cancer immunity
EMBO Molecular Medicine (2023)
-
The role of lysosomes in metabolic and autoimmune diseases
Nature Reviews Nephrology (2023)
-
Dissolution of oncofusion transcription factor condensates for cancer therapy
Nature Chemical Biology (2023)
-
The Roles of Autophagy in the Genesis and Development of Polycystic Ovary Syndrome
Reproductive Sciences (2023)