Abstract
The transcriptional coactivator Yes-associated protein 1 (YAP) orchestrates a proproliferative transcriptional program that controls the fate of somatic stem cells and the regenerative responses of certain tissues. As such, agents that activate YAP may hold therapeutic potential in disease states exacerbated by insufficient proliferative repair. Here we report the discovery of a small molecule, termed PY-60, which robustly activates YAP transcriptional activity in vitro and promotes YAP-dependent expansion of epidermal keratinocytes in mouse following topical drug administration. Chemical proteomics revealed the relevant target of PY-60 to be annexin A2 (ANXA2), a protein that directly associates with YAP at the cell membrane in response to increased cell density. PY-60 treatment liberates ANXA2 from the membrane, ultimately promoting a phosphatase-bound, nonphosphorylated and transcriptionally active form of YAP. This work reveals ANXA2 as a previously undescribed, druggable component of the Hippo pathway and suggests a mechanistic rationale to promote regenerative repair in disease.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA-seq data that support the findings of this study have been deposited in GEO with the accession code GSE164801. Source data are provided with this paper.
References
Justice, R. W., Zilian, O., Woods, D. F., Noll, M. & Bryant, P. J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546 (1995).
Xu, T., Wang, W., Zhang, S., Stewart, R. A. & Yu, W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 (1995).
Yu, F. X., Zhao, B. & Guan, K. L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 (2015).
Gumbiner, B. M. & Kim, N. G. The Hippo–YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 127, 709–717 (2014).
Yu, F. X. & Guan, K. L. The Hippo pathway: regulators and regulations. Genes Dev. 27, 355–371 (2013).
Meng, Z., Moroishi, T. & Guan, K. L. Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016).
Meng, Z. et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 6, 8357 (2015).
Johnson, R. & Halder, G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 13, 63–79 (2014).
Lian, I. et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 24, 1106–1118 (2010).
Lavado, A. et al. The hippo pathway prevents YAP/TAZ-driven hypertranscription and controls neural progenitor number. Dev. Cell 47, 576–591 (2018).
Yui, S. et al. YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22, 35–49 (2018).
Gregorieff, A., Liu, Y., Inanlou, M. R., Khomchuk, Y. & Wrana, J. L. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526, 715–718 (2015).
Schlegelmilch, K. et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 144, 782–795 (2011).
Hong, A. W., Meng, Z. & Guan, K. L. The Hippo pathway in intestinal regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 13, 324–337 (2016).
Panciera, T. et al. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell 19, 725–737 (2016).
Leach, J. P. et al. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550, 260–264 (2017).
Fan, F. et al. Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration. Sci. Transl. Med. 8, 352ra108 (2016).
Galan, J. A. & Avruch, J. MST1/MST2 protein kinases: regulation and physiologic roles. Biochemistry 55, 5507–5519 (2016).
Roh, K. H. & Choi, E. J. TRAF2 functions as an activator switch in the reactive oxygen species-induced stimulation of MST1. Free Radic. Biol. Med. 91, 105–113 (2016).
Chae, J. S., Gil Hwang, S., Lim, D. S. & Choi, E. J. Thioredoxin-1 functions as a molecular switch regulating the oxidative stress-induced activation of MST1. Free Radic. Biol. Med. 53, 2335–2343 (2012).
Hergovich, A. & Hemmings, B. A. Hippo signalling in the G2/M cell cycle phase: lessons learned from the yeast MEN and SIN pathways. Semin. Cell Dev. Biol. 23, 794–802 (2012).
Praskova, M., Xia, F. & Avruch, J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18, 311–321 (2008).
Lehtinen, M. K. et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125, 987–1001 (2006).
DeRan, M. et al. Energy stress regulates Hippo–YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein. Cell Rep. 9, 495–503 (2014).
Miller, E. et al. Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP. Chem. Biol. 19, 955–962 (2012).
Zhang, N. et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27–38 (2010).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Cordenonsi, M. et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 (2011).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Overholtzer, M. et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl Acad. Sci. USA 103, 12405–12410 (2006).
Zhao, B. et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 (2011).
Totaro, A. et al. YAP/TAZ link cell mechanics to notch signalling to control epidermal stem cell fate. Nat. Commun. 8, 15206 (2017).
Pepe-Mooney, B. J. et al. Single-cell analysis of the liver epithelium reveals dynamic heterogeneity and an essential role for YAP in homeostasis and regeneration. Cell Stem Cell 25, 23–38 (2019).
Rescher, U. & Gerke, V. Annexins—unique membrane binding proteins with diverse functions. J. Cell Sci. 117, 2631–2639 (2004).
Lee, D. B., Jamgotchian, N., Allen, S. G., Kan, F. W. & Hale, I. L. Annexin A2 heterotetramer: role in tight junction assembly. Am. J. Physiol. Renal Physiol. 287, F481–F491 (2004).
Heyraud, S. et al. Contribution of annexin 2 to the architecture of mature endothelial adherens junctions. Mol. Cell. Biol. 28, 1657–1668 (2008).
Hauri, S. et al. Interaction proteome of human Hippo signaling: modular control of the co-activator YAP1. Mol. Syst. Biol. 9, 713 (2013).
Benaud, C. et al. AHNAK interaction with the annexin 2/S100A10 complex regulates cell membrane cytoarchitecture. J. Cell Biol. 164, 133–144 (2004).
Rowan, M. P. et al. Burn wound healing and treatment: review and advancements. Crit. Care 19, 243 (2015).
Han, G. & Ceilley, R. Chronic wound healing: a review of current management and treatments. Adv. Ther. 34, 599–610 (2017).
Yin, F. et al. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154, 1342–1355 (2013).
Ling, Q. et al. Annexin II regulates fibrin homeostasis and neoangiogenesis in vivo. J. Clin. Invest. 113, 38–48 (2004).
Acknowledgements
This work was supported by the Skaggs Institute for Chemical Biology and Calibr. We thank A. Davila and members of the compound management group at Calibr for excellent technical assistance.
Author information
Authors and Affiliations
Contributions
S.Z.S. performed mouse experiments, isothermal titration calorimetry, biolayer interferometry experiments and primary keratinocyte proliferation experiments. P.-Y.Y. performed pilot screens, identified hit compounds, synthesized chemical analogs and designed and identified PY-60 and PY-PAP. E.M.G. expressed all recombinant proteins and performed reporter assays. K.N. performed fluorescence polarization experiments. S.S. performed proliferation assays and gene expression experiments. C.Z. synthesized the PY-60 and PY-PAP used in this work. J.I. and S.R.C. constructed and prepared expression vectors. P.-Y.Y., E.C., S.L. and M.H. performed the high-throughput chemical screens and validated hit compounds. L.I. retrieved historical expression data from skin. C.S. performed docking studies. M.J.B. carried out the target identification work, performed imaging, did in situ target engagement work, performed RNA-seq, did immunoprecipitation experiments, performed RT–qPCR experiments and performed all immunoblotting. S.Z.S., P.-Y.Y., E.M.G., K.N., S.S., L.I., C.S., E.C., S.L., M.H. and M.J.B. analyzed primary data. P.-Y.Y., W.S., P.G.S. and M.J.B. conceived the project idea. M.D. and X.W. contributed the reporter cell line and experimental advice on evaluation of expression data. A.K.C and P.-Y.Y. designed chemical analogs. A.K.C., W.S., F.D.C., P.G.S. and M.J.B. supervised the work. S.Z.S., F.D.C., P.G.S. and M.J.B. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
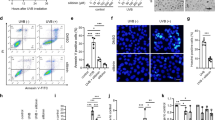
Extended Data Fig. 1 PY-60 activates a robust, YAP- dependent transcriptional program in multiple cell types.
a, 14xTEAD-LUC assay, comparing PY-60 treatment (10 µM) to concomitant siRNA-mediated NF2/LATS1/2 knockdown (n=3, mean and s.d.). b, BRET ratio of the TEAD-YAP interaction (mBU = BRET units; n=3, mean and s.d.), comparing PY-60 treatment (10 µM) to NF2/LATS1/2 siRNA treatment. c, Luminance values from a split luciferase assay measuring the association of between YAP and TEAD in the presence of PY-60 (n=3, mean and s.d.) d, Nuclear to cytoplasmic ratios of anti- YAP immunofluorescence intensities from high content image analysis from MDCK cells at the indicated cell densities (cells per well in 384-well plates) for 48 hours with PY-60 (10 µM; mean and s.d.). e-h, Transcript levels of YAP-controlled genes from human cell lines treated with PY-60 for 24 hours (n=3, mean and s.d.). i, TEAD- LUC activity in response to PY-60 treatment (n=3; mean and s.e.m.) and Western blots of YAP knockdown from 293A cells with stably integrated shRNAs that are non- targeting (scramble) or that target YAP1. j, Transcript levels of YAP dependent genes or YAP1 itself and anti- YAP Western blot analysis in response to PY-60 treatment (10 µM; n=3, mean and s.d.; perecentage indicative of transcript knockdown) in 293A cells with stable shRNAs that are non-targeting (scramble) or targeting YAP1 (univariate two-sided t-test; NS= not statistically significant). Exeriments are mean and s.d. or s.e.m. as noted of biologically independent samples. Western blots (i, j) are representative of experimental replicates (n = 2).
Extended Data Fig. 2 PY-60 promotes anchorage- and contact inhibition- independent growth of epithelial cell lines.
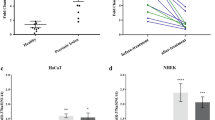
a, Quantification and representative images of MCF10A colonies formed in soft agar after 2-week treatment with PY-60 (n=3, mean and s.e.m.,univariate two sided t-test; scale bar = 100 µm). b, Representative confocal images (n = 4) of MDCK cells treated with DMSO or PY-60 (10 µM) for 10 days and stained with Hoechst 33342 (DNA) and Phalloidin-488 (F-actin), depicting the loss cell contact inhibition of growth (scale bar = 5 µm). c, Relative transcript levels of Yap1 and Taz from mouse skin, epidermis, and primary keratinocytes from publicly available RNA-seq data accessed from the indicated GEO database entries (n=3 biologically ditinct samples; mean and s.e.m.).
Extended Data Fig. 3 PY-60 promotes YAP dephosphorylation and activates a more robust YAP-dependent transcriptional program than MST1/2 inhibitor XMU-MP-1.
a, Representative (n=3) Western blot analysis of P-YAP (S127) and total YAP levels from 293A cells treated with the indicated compounds for 24 hours. b, Relative TEAD- LUC luminance values from 293A-TEAD-LUC cells treated for 24 hours with PY-60 or XMU-MP-1 (n=3, mean and s.e.m.) c,d, Transcript levels of YAP target genes ANKRD1 and CYR61 from 293A cells treated for 24 hours with PY-60 and XMU-MP-1 (n=3, mean and s.d.). e,f, Quantification and representative images of nuclei from 293A cells treated with PY-60 (20 µM) or XMU-MP-1 (1 µM) for 24 hours (n =3 biologically indepent samples, mean and s.d.; scale bar = 5 µm). Statistical tests are two sided univariate t-tests. Data are mean and s.d. or s.e.m. as noted of biologically independent samples.
Extended Data Fig. 4 Chemical proteomics identifies ANXA2 as a druggable component of the Hippo pathway.
a, Activities of the indicated Hippo pathway kinases in the presence of PY-60 and positive control inhibitors XMU- MP-1 (MST1/2) and Staurosporine (STS; LATS1/2, n=3; mean and s.e.m.). b, Represenative fluorescent gel scans of rhodamine-azide labeled 293A cell lysates after in situ crosslinking with PY-PAP (n=2). c, Anti-biotin Western blot and silver stained gel of streptavidin-enriched material after in situ crosslinking with PY-PAP (10 µM) in 293A cells (n=4). d, Anti-biotin and anti-ANXA2 Western blot analyses of streptavidin-enriched material after in situ treatment of 293A cells with PY-PAP (10 µM) or excess PY-60 (100 µM). Arrows indicate the competitively labeled band corresponding to ANXA2 and the band not competed by PY-60 corresponding to CTSD (n=2). e, Summary of MS/MS results, depicting the identified tryptic peptides and Metlin scores of ANXA2 (n=5). f, Isothermal titration calorimetry analysis of ANXA2 association with PY-60 (Kd = 22 µM; n=3) g, h, Quantification and representative Western blotting analysis of ANXA2 levels in the presence of PY-60 (20 µM) in cellular thermal shift assays with 293A cells (n=3; mean and s.d.). i, Dye-based thermal denaturation assay with recombinant ANXA2 protein in the presence of PY-60 (20 µM) or DMSO (n = 3; mean and s.d.). j, Quantification of ANXA2 transcript knockdown in 293A cells subjected to the indicated siRNAs. k, Western blot analyses of P-YAP levels in response to siRNA mediated knockdown of ANXA2 (n=2). l, Relative ANXA2 transcript levels from 293A cells stably lentivirally transduced with the indicated ANXA2-targeted shRNAs as in Fig. 3h. m, Relative transcript levels of YAP-driven transcript of CYR61 in response to stable knockdown of CTSD or ANXA2 in 293A cells with respective quantification of target transcript knockdown (middle, right; n =3; mean and s.d.). n, Relative ANXA2 levels from 293A cells stably expressing ANXA2-targeted shRNA as in Fig. 3i. o, Western blot analyses of ANXA2 levels from 293A and 293A-TEAD-LUC cells stably overexpressing ANXA2 as in Fig. 3h,i (n=2). Statistical tests are two sided univariate t-tests. Data are mean and s.d. or s.e.m. as noted of biologically independent samples.
Extended Data Fig. 5 Loss of ANXA2 promotes proliferation in growth-limiting conditions.
a,b, Number of colonies and Western blotting of ANXA2 levels with quantification of protein levels from MCF10A cells transduced with ANXA2 targeted shRNAs (n = 3; mean and s.d.). c,d, Number of colonies and representative anti-FLAG Western blot of MCF10A cells in soft agar after transduction with lentiviruses encoding a vector or ANXA2-FLAG transgene and treated with PY-60 (n = 6; mean and s.d). e,f, Quantification of cell density and representative Western blotting for ANXA2 levels from MDCK cells stably expressing ANXA2-targeted shRNAs (n = 3; mean and s.d.). g,h, Quantification of cell density and representative Western blotting for ANXA2 levels from MDCK cells stably overexpressing vector or ANXA2-FLAG transgene and exposed to PY-60 for 7 days (n = 3; mean and s.d.; t test). i-k, Number of cells, representative rhodamine B staining, and anti-ANXA2 Western blotting of human keratinocytes (HaCaT) stably expressing the indicated shRNAs (0.5% FBS, 7 days; n = 3; mean and s.d.; scale bar = 7 mm). l-n, Number of cells, representative Rhodamine B staining (2% FBS), and anti- FLAG Western blotting of human keratinocytes stably overexpressing vector or ANXA2-FLAG transgene and treated with PY-60 (1 µM; 7 days; n = 3; mean and s.d.; t test; scale bar = 7 mm). Statistical tests are two sided univariate t-tests. Data are mean and s.d. or s.e.m. as noted of biologically independent samples.
Extended Data Fig. 6 Hippo pathway member ANXA2 binds to MST2.
a, Western blotting analysis for HA-MST2 from anti-FLAG immunoprecipitated material from confluent 293A cells transiently overexpressing MST2-HA and ANXA2-FLAG treated with DMSO or PY-60 (20 µM; n=2). b, Western blotting for MST1-HA or MST2-HA from anti-FLAG immunoprecipitated material from confluent 293A cells transiently overexpressing MST1-HA or MST2-HA and ANXA2-FLAG. c,d, Schematic and Western blotting analysis of HA-tagged transgenes after anti-FLAG immunoprecipitation from 293A cells expressing MST1/2 domain swapped proteins and ANXA2-FLAG (ID = inhibitory domain; SARAH = Sav-RASSF-Hpo SARAH domain; n=3).e, Biolayer interferometry-based quantification of the dissociation constant of MST2 with ANXA2 (n=3 biologically independent samples; mean and s.e.m.). Western blotting is representative of independent experimental replicates as noted.
Extended Data Fig. 7 ANXA2 binds YAP and MST2, an interaction not competed by PY- 60.
a, Western blotting for endogenous YAP from anti-FLAG immunoprecipitated material from 293A cells transiently overexpressing ANXA2-FLAG then subjected to chemical crosslinker DSS. b, Western blotting analysis of endogenous MST2 prtein from anti-FLAG immunoprecipitation from 293A cells transiently overexpressing ANXA2-FLAG then subjected to crosslinker DSS (0.1-5 mM). c, Western blotting analysis of endogenous YAP from anti-FLAG immunoprecipitated material from 293A cells transiently overexpressing ANXA2-FLAG subjected to PY-60 for 24 hours and then treated with crosslinker DSS (1 mM). d, Western blotting of endogenous MST2 from anti-FLAG immunoprecipitated material from 293A cells transiently overexpressing ANXA2-FLAG treated with PY-60 for 24 hours and then exposed to DSS (1 mM). Western blotting are representative of biologically distinct experimental replicates (n=3).
Extended Data Fig. 8 ANXA2 binds to YAP and TAZ, an interaction not competed by PY-60 treatment.
a, Biolayer interferometry analysis demonstrating no change in interaction between ANXA2 and YAP in the presence of the indicated concentration response of PY-60 (n=3; mean and s.e.m of biologically independent replicates). b, Western blotting analysis for the presence of HA-tagged TAZ of anti-FLAG precipitated material from 293A cells transiently transfected with ANXA2-FLAG and HA-TAZ with PY-60 treatment (20 µM) for 24 hours. c, Western blotting analysis for anti-phospho-TAZ content from 293A cells treated for 24 hours with the indicated compounds. Western blots are representative of biologically distinct experimental replicates (n=2).
Extended Data Fig. 9 Characterization of PY-60 binding to ANXA2, a phosphoinositide associated membrane protein.
a, Colocalization coefficients of ANXA2 and pan-cadherin co-staining from 293A cells treated with PY-60 as depicted in Fig. 6a (n=3). b,c, Schematic and representative anti-ANXA2 Western blotting analysis of recombinant ANXA2 binding to the phosphoinositide membrane array (PIP strips, Echelon Biosciences; LPA = lysophosphatidic acid; LPC= lysophosphatidylcholine; PtdIns = phosphatidylinositol; PE = phosphoethanolamine; PA = phosphatidic acid; PS = phosphatidylserine). d, Quantification of ANXA2 binding to phosphoinositides in the PIP strip array (n=3). e, Structure of BODIPY fluorophore conjugated phosphoinositides, PtdIns-BODIPY and PtdIns-(4,5)P2-BODIPY. f,g, Fluorescent scan and quantification of Rhodamine azide (Rh-N3) labeled ANXA2-FLAG following immunoprecipitation after in situ crosslinking with PY-PAP (n=3). h, Schematic depicting the FLAG- tagged annexin repeat transgenes used. i, Fluorescent scan and anti-FLAG Western blot of Rhodamine azide (Rh-N3) labeled ANXA2-FLAG transgenes following anti-FLAG immunoprecipitation after in situ treatment with PY-PAP and UV crosslinking. j, Docked pose of PY-60 and PY-PAP to ANXA2 (reference structure PDB ID: 1XJL) with annexin repeats color coded. k, Isolated pose of PY-60 and PY-PAP docked to the first repeat of ANXA2 with residues deemed essential or dispensable by in situ labeling studies indicated in magenta and blue respectively. l,m, Fluorescent scans and quantification of Rhodamine azide labeled ANXA2-FLAG after anti-FLAG precipitation from 293A cells treated in situ with PY-PAP (10 µM) and crosslinked (n=3). Data reported are mean and s.d. of biologically independent samples. Statistical tests are two sided univariate t-tests.
Extended Data Fig. 10 PPP2CA is essential for YAP activation by PY-60.
a, Representative biolayer interferometry curve (n=4 independent experimental replicates) demonstrating the association of PPP2CA with ANXA2. b, Representative anti-PPP2CA Western blotting analysis (n=2) from stable 293A-TEAD-LUC reporter cells harboring the indicated shRNAs. c, Relative transcript levels of YAP-dependent transcripts from 293A cells treated with PY-60 (10 µM) for 24 hours (n=3 biologically independent samples, mean and s.d.). d, Representative Western blotting anlaysis (n=2) from 293A cell lines stably expressing the indicated PPP2CA shRNAs. Western blots are representative of independent experimental replicates as noted.
Supplementary information
Supplementary Information
Supplementary Note 1 and Tables 1–4.
Source data
Source Data Fig. 3
Unprocessed immunoblots and gels.
Source Data Fig. 4
Unprocessed immunoblots and gels.
Source Data Fig. 5
Unprocessed immunoblots and gels.
Source Data Fig. 6
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 1
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 3
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 4
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 5
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 6
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 7
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 8
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 9
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 10
Unprocessed immunoblots and gels.
Rights and permissions
About this article
Cite this article
Shalhout, S.Z., Yang, PY., Grzelak, E.M. et al. YAP-dependent proliferation by a small molecule targeting annexin A2. Nat Chem Biol 17, 767–775 (2021). https://doi.org/10.1038/s41589-021-00755-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00755-0
This article is cited by
-
PIN1 promotes the metastasis of cholangiocarcinoma cells by RACK1-mediated phosphorylation of ANXA2
Cellular Oncology (2024)
-
SEC23A confers ER stress resistance in gastric cancer by forming the ER stress-SEC23A-autophagy negative feedback loop
Journal of Experimental & Clinical Cancer Research (2023)
-
YAP-galectin-3 signaling mediates endothelial dysfunction in angiotensin II-induced hypertension in mice
Cellular and Molecular Life Sciences (2023)
-
The Hippo signalling pathway and its implications in human health and diseases
Signal Transduction and Targeted Therapy (2022)
-
Effect of cell density on the malignant biological behavior of breast cancer by altering the subcellular localization of ANXA2 and its clinical implications
Clinical and Translational Oncology (2022)