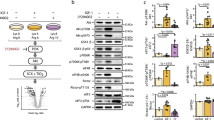
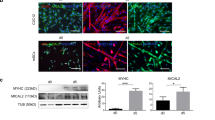
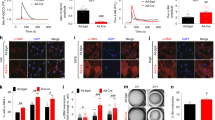
Abstract
Many intracellular proteins are modified by N-acetylglucosamine, a post-translational modification termed O-GlcNAc. This modification is found on serine and threonine side chains and has the potential to regulate signaling pathways through interplay with phosphorylation. Here, we discover and characterize one such example. We find that O-GlcNAc levels control the sensitivity of fibroblasts to actin contraction induced by the signaling lipid sphingosine-1-phosphate (S1P), culminating in the phosphorylation of myosin light chain (MLC) and cellular contraction. Specifically, O-GlcNAc modification of the phosphatase subunit MYPT1 inhibits this pathway by blocking MYPT1 phosphorylation, maintaining its activity and causing the dephosphorylation of MLC. Finally, we demonstrate that O-GlcNAc levels alter the sensitivity of primary human dermal fibroblasts in a collagen-matrix model of wound healing. Our findings have important implications for the role of O-GlcNAc in fibroblast motility and differentiation, particularly in diabetic wound healing.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RT-PCR primers were designed using the MIT Primer3 design database (http://biotools.nubic.northwestern.edu/Primer3.html) with sequences found using the NCBI Gene Search tool (ncbi.nlm.nih.gov/gene). Source data are provided with this paper.
References
Yang, X. & Qian, K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465 (2017).
Zachara, N. E. Critical observations that shaped our understanding of the function(s) of intracellular glycosylation (O‐Glc NAc). FEBS Lett. 592, 3950–3975 (2018).
King, D. T., Males, A., Davies, G. J. & Vocadlo, D. J. Molecular mechanisms regulating O-linked N-acetylglucosamine (O-GlcNAc)-processing enzymes. Curr. Opin. Chem. Biol. 53, 131–144 (2019).
Hart, G. W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 294, 2211–2231 (2019).
Marshall, S., Bacote, V. & Traxinger, R. R. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. ole of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem. 266, 4706–4712 (1991).
Shen, D. L., Gloster, T. M., Yuzwa, S. A. & Vocadlo, D. J. Insights into O-linked N-acetylglucosamine (O-GlcNAc) processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates. J. Biol. Chem. 287, 15395–15408 (2012).
Hart, G. W., Slawson, C., Ramirez-Correa, G. & Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription and chronic disease. Annu. Rev. Biochem. 80, 825–858 (2011).
Wang, S. et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates Akt signaling. PLoS ONE 7, e37427 (2012).
Tarrant, M. K. et al. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat. Chem. Biol. 8, 262–269 (2012).
Gloster, T. M. et al. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat. Chem. Biol. 7, 174–181 (2011).
Jiang, H., Rhee, S., Ho, C.-H. & Grinnell, F. Distinguishing fibroblast promigratory and procontractile growth factor environments in 3-D collagen matrices. FASEB J. 22, 2151–2160 (2008).
Yuzwa, S. A. et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490 (2008).
Spiegel, S. & Milstien, S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4, 397–407 (2003).
Watterson, K. R., Lanning, D. A., Diegelmann, R. F. & Spiegel, S. Regulation of fibroblast functions by lysophospholipid mediators: potential roles in wound healing. Wound Repair Regen. 15, 607–616 (2007).
Takabe, K., Paugh, S. W., Milstien, S. & Spiegel, S. ‘Inside-out’ signaling of sphingosine-1-phosphate: therapeutic targets. Pharmacol. Rev. 60, 181–195 (2008).
Mendelson, K., Evans, T. & Hla, T. Sphingosine 1-phosphate signalling. Development 141, 5–9 (2013).
Eto, M. & Kitazawa, T. Diversity and plasticity in signaling pathways that regulate smooth muscle responsiveness: paradigms and paradoxes for the myosin phosphatase, the master regulator of smooth muscle contraction. J. Smooth Muscle Res. 53, 1–19 (2017).
Rhee, S. & Grinnell, F. Fibroblast mechanics in 3D collagen matrices. Adv. Drug Deliv. Rev. 59, 1299–1305 (2007).
Riento, K. & Ridley, A. J. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 4, 446–456 (2003).
Amano, M., Nakayama, M. & Kaibuchi, K. Rho-kinase/ROCK: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton 67, 545–554 (2010).
Totsukawa, G. et al. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell Biol. 150, 797–806 (2000).
Feng, J. et al. Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J. Biol. Chem. 274, 37385–37390 (1999).
Kawano, Y. et al. Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J. Cell Biol. 147, 1023–1038 (1999).
Murányi, A. et al. Phosphorylation of Thr695 and Thr850 on the myosin phosphatase target subunit: inhibitory effects and occurrence in A7r5 cells. FEBS Lett. 579, 6611–6615 (2005).
Khasnis, M., Nakatomi, A., Gumpper, K. & Eto, M. Reconstituted human myosin light chain phosphatase reveals distinct roles of two inhibitory phosphorylation sites of the regulatory subunit, MYPT1. Biochemistry 53, 2701–2709 (2014).
Heissler, S. M. & Sellers, J. R. Myosin light chains: teaching old dogs new tricks. Bioarchitecture 4, 169–188 (2014).
Ito, M., Nakano, T., Erdodi, F. & Hartshorne, D. J. Myosin phosphatase: structure, regulation and function. Mol. Cell. Biochem. 259, 197–209 (2004).
Grassie, M. E., Moffat, L. D., Walsh, M. P. & MacDonald, J. A. The myosin phosphatase targeting protein (MYPT) family: a regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1δ. Arch. Biochem. Biophys. 510, 147–159 (2011).
Takemoto, K., Ishihara, S., Mizutani, T., Kawabata, K. & Haga, H. Compressive stress induces dephosphorylation of the myosin regulatory light chain via RhoA phosphorylation by the adenylyl cyclase/protein kinase A signaling pathway. PLoS ONE 10, e0117937 (2015).
Hédou, J. et al. O-linked N-acetylglucosaminylation is involved in the Ca2+ activation properties of rat skeletal muscle. J. Biol. Chem. 282, 10360–10369 (2007).
Cheung, W. D., Sakabe, K., Housley, M. P., Dias, W. B. & Hart, G. W. O-linked β-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem. 283, 33935–33941 (2008).
Clark, P. M. et al. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J. Am. Chem. Soc. 130, 11576–11577 (2008).
Wang, S. et al. Quantitative proteomics identifies altered O-GlcNAcylation of structural, synaptic and memory-associated proteins in Alzheimer’s disease. J. Pathol. 243, 78–88 (2017).
Qin, K. et al. Quantitative profiling of protein O-GlcNAcylation sites by an isotope-tagged cleavable linker. ACS Chem. Biol. 13, 1983–1989 (2018).
Li, J. et al. An isotope-coded photocleavable probe for quantitative profiling of protein O-GlcNAcylation. ACS Chem. Biol. 14, 4–10 (2019).
Ichikawa, K. et al. Interactions and properties of smooth muscle myosin phosphatase. Biochemistry 35, 6313–6320 (1996).
Hirano, K., Phan, B. C. & Hartshorne, D. J. Interactions of the subunits of smooth muscle myosin phosphatase. J. Biol. Chem. 272, 3683–3688 (1997).
Tanaka, J. et al. Interaction of myosin phosphatase target subunit 1 with the catalytic subunit of type 1 protein phosphatase. Biochemistry 37, 16697–16703 (1998).
Wang, Y. et al. ROCK isoform regulation of myosin phosphatase and contractility in vascular smooth muscle cells. Circ. Res. 104, 531–540 (2009).
Rexach, J. E. et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol. 6, 645–651 (2010).
Darabedian, N., Thompson, J. W., Chuh, K. N., Hsieh-Wilson, L. C. & Pratt, M. R. Optimization of chemoenzymatic mass tagging by strain-promoted cycloaddition (SPAAC) for the determination of O-GlcNAc stoichiometry by western blotting. Biochemistry 57, 5769–5774 (2018).
Gosain, A. & DiPietro, L. A. Aging and wound healing. World J. Surg. 28, 321–326 (2004).
Guo, S. & DiPietro, L. A. Factors affecting wound healing. J. Dent. Res. 89, 219–229 (2010).
Grinnell, F. & Petroll, W. M. Cell motility and mechanics in three-dimensional collagen matrices. Annu. Rev. Cell Dev. Biol. 26, 335–361 (2010).
Kawanabe, T., Kawakami, T., Yatomi, Y., Shimada, S. & Soma, Y. Sphingosine 1-phosphate accelerates wound healing in diabetic mice. J. Dermatol. Sci. 48, 53–60 (2007).
Aoki, M. et al. Sphingosine-1-phosphate facilitates skin wound healing by increasing angiogenesis and inflammatory cell recruitment with less scar formation. Int. J. Mol. Med. 20, E3381 (2019).
Somlyo, A. P. & Somlyo, A. V. Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases and myosin phosphatase. Physiol. Rev. 83, 1325–1358 (2003).
Grassie, M. E. et al. Cross-talk between Rho-associated kinase and cyclic nucleotide-dependent kinase signaling pathways in the regulation of smooth muscle myosin light chain phosphatase. J. Biol. Chem. 287, 36356–36369 (2012).
Qiao, Y.-N. et al. Myosin phosphatase target subunit 1 (MYPT1) regulates the contraction and relaxation of vascular smooth muscle and maintains blood pressure. J. Biol. Chem. 289, 22512–22523 (2014).
Liu, C. et al. O-GlcNAcylation of myosin phosphatase targeting subunit 1 (MYPT1) dictates timely disjunction of centrosomes. J. Biol. Chem. 295, 7341–7349 (2020).
Acknowledgements
We thank F. Grinnell for consultations on collagen matrix contraction. This research was supported by the American Cancer Society Research Scholar Grant (RSG-14-225-01-CCG), the University of Southern California, the Anton Burg Foundation and the National Institutes of Health (R01GM125939; to M.R.P.). N.J.P. is supported by NIGMS T32GM118289. We thank K. Moremen at the University of Georgia, who is supported by the National Institutes of Health (P41GM103390 and R01GM130915), for the generous gift of GalT(Y289L).
Author information
Authors and Affiliations
Contributions
N.J.P., A.R.B., N.D. and M.R.P. designed experiments and interpreted data. N.J.P. and A.R.B. carried out cellular phenotype and western blotting experiments. N.J.P. generated the MYPT1 stable cell lines and performed the associated experiments. N.J.P. also performed the co-IP, apoptosis and collagen matrix experiments. N.D. performed the analysis of MYPT1 O-GlcNAc levels and dynamics. N.J.P., A.R.B. and M.R.P. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Figure 1 Modulating O-GlcNAcylation levels and identification of S1P.
a, Schematic layout of our general experimental conditions. b, O-GlcNAc levels can be changed upon OGT or OGA inhibition. NIH3T3 cells were treated with either the OGT inhibitor 5SGlcNAc, the OGA inhibitor Thiamet G, or DMSO vehicle respectively before the O-GlcNAc levels were analyzed by Western blotting. c, Only serum- or S1P-treatment results in notable cell contraction when O-GlcNAc levels have been lowered. NIH3T3 cells were treated with DMSO or 5SGlcNAc. The indicated signaling molecules were then added, and the contraction phenotype was then visualized using bright-field microscopy. The data in b-c is representative of 2 biological replicates.
Extended Data Figure 2 O-GlcNAc controls the sensitivity of fibroblasts to sphingosine-1-phosphate (S1P) mediated cell contraction.
a, O-GlcNAc levels can be changed upon OGT inhibition. NIH3T3 cells were treated with either the OGT inhibitor ST060266 or DMSO vehicle respectively before the O-GlcNAc levels were analyzed by Western blotting. b, O-GlcNAc levels can be lowered by OGT RNAi. NIH3T3 cells were transfected with RNAi targeting OGT or a scrambled sequence before analysis by Western blotting. c, NIH3T3 cells were treated with the indicated combinations of DMSO, the OGT inhibitor ST060266 (200 μM) and/or S1P. The contraction phenotype was then visualized using bright-field microscopy. d, NIH3T3 cells were transfected with either scramble or OGT-targeted RNAi for 48 h before addition of either DMSO or S1P. The contraction phenotype was then visualized using bright-field microscopy. e, O-GlcNAc levels can be changed by culturing cells in different glucose concentrations. NIH3T3 cells were cultured in the indicated concentrations of glucose before analysis by Western blotting. The data in a-e is representative of at least 2 biological replicates.
Extended Data Figure 3 Signaling through the second S1P receptor, S1PR2, is responsible for the contraction phenotype.
a, NIH3T3 cells can express all five S1P GPCRs (S1PR1 to 5). mRNA was collected from NIH3T3 cells before being subjected to RT-PCR and visualization on an DNA-agarose gel. These data are representative of 2 biological replicates. b, Antagonizing S1PR2, but not the other receptors, inhibits S1P-mediated cell contraction. NIH3T3 cells were treated with either DMSO or 5SGlcNAc. The same cells were then treated with either additional DMSO or the indicated selective antagonists followed by S1P. The contraction phenotype was then visualized using bright-field microscopy. c, Quantitation of the data in (b). Results are the mean ± SEM of the relative culture plate area taken-up by cells in at least three randomly selected frames. Statistical significance was determined using a 2-tailed student’s t-test. d, S1PR5 agonism does not induce cell contraction. NIH3T3 cells were treated with either DMSO or 5SGlcNAc. The same cells were then treated with either S1P or the S1PR5-selective agonist A971432. The contraction phenotype was then visualized using bright-field microscopy. e, Quantitation of the data in (d). Results are the mean ± SEM of the relative culture plate area taken-up by cells in at least three randomly selected frames. Statistical significance was determined using a 2-tailed student’s t-test. f, Lowering O-GlcNAc levels increases the sensitivity of NIH3T3 cells to S1PR2 induced cell contraction. NIH3T3 cells were treated with either DMSO or 5SGlcNAc before addition of the indicated concentrations of the S1PR2-selective agonist CYM5220. The contraction phenotype was then visualized using bright-field microscopy. g, Quantitation of the data in (f). Results are the mean ± SEM of the relative culture plate area taken-up by cells in four randomly selected frames. Statistical significance was determined using a 2-way ANOVA test followed by Sidak’s multiple comparisons test. h, S1PR2 knockdown using siRNA blocks contraction phenotype. NIH3T3 cells were transfected with either scramble or S1Pr2-targeted RNAi before addition of DMSO or S1P. The contraction phenotype was then visualized using bright-field microscopy. i, Quantitation of the data in (h). Results are the mean ± SEM of the relative culture plate area taken-up by cells in three randomly selected frames. Statistical significance was determined using a 2-way ANOVA test followed by Sidak’s multiple comparisons test.
Extended Data Figure 4 Inhibition of Rho kinase (ROCK1/2) blocks S1P-mediated cell contraction.
NIH3T3 cells were treated with either DMSO or 5SGlcNAc. The same cells were then treated with either additional DMSO or the ROCK1/2 inhibitor Y27632 followed by the indicated concentrations of S1P. The contraction phenotype was then visualized using bright-field microscopy. The results were then quantified and are presented as mean ± SEM of the relative culture plate area taken-up by cells in four randomly selected frames.
Extended Data Figure 5 Analysis of MYPT1 mutants shows that deletion of the serine/threonine domain sensitizes cells to S1P-mediated contraction.
a, Sequence alignment of MYPT1 and the different mutants tested here. Red indicates potential O-GlcNAc modification sites (S or T), and blue indicates O-GlcNAc sites previously identified by mass spectrometry. MYTP1(S/TtoA) mutates all of the previously identified O-GlcNAc sites, MYPT1(d564-578) deletes the first serine/threonine rich region, MYPT1(d588-602) deletes the second serine/threonine rich region, and MYPT1Δ deletes the entire serine/threonine rich domain. b, MYPT1Δ, and to a lesser extent MYPT1(d564-578), sensitizes cells to S1P-mediated contraction. NIH3T3 cells expressing the indicated MYPT1 proteins and the endogenous copy was removed by RNAi. DMSO or S1P was then added and the contraction of the cells was measured. Results are the mean ± SEM of the relative culture plate area taken-up by cells in four randomly selected frames. Statistical significance was determined using a 2-way ANOVA test followed by Sidak’s multiple comparisons test.
Extended Data Figure 6 Deletion of the MYPT1 serine/threonine rich domain yields MYPT1Δ with reduced O-GlcNAc modification.
a, Schematic of the MYPT1Δ protein, which lacks the major O-GlcNAc region of the protein. b&c) MYPT1Δ loses a notable amount of O-GlcNAc. The O-GlcNAc levels of FLAG-tagged MYPT1 or MYPT1Δ were analyzed using mass shifting or IP-Western blot. The data in are representative of at least 2 biological replicates.
Extended Data Figure 7 MYPT1Δ is an active phosphatase that can dephosphorylate MLC and return cells to a relaxed state.
a, If MYPT1Δ is an active phosphatase, we expect that it will become phosphorylated and deactivated by ROCK after S1P treatment but will return to a desphosphorylated and active state after a longer period of time. This will result in MLC dephosphorylation and relaxation of the cells. This is exactly what we observed by Western blotting. These data are representative of 2 biological replicates. b, Cells expressing MYPT1Δ return to a relaxed state over 180 min. The contraction phenotype was visualized using bright-field microscopy. The results were then quantified and are presented as mean ± SEM of the relative culture plate area taken-up by cells in four randomly selected frames. Statistical significance was determined using a 2-way ANOVA test followed by Sidak’s multiple comparisons test.
Extended Data Figure 8 O-GlcNAc blocks the interaction between MYPT1 and ROCK.
NIH3T3 cells expressing flag-tagged MYPT1 were treated with either DMSO or 5SGlcNAc (200 μM). An anti-flag co-immunoprecipitation was then performed using the Catch and Release system (Thermo). ROCK enrichment was detected by Western blotting and normalized to overall protein capture (Coomassie staining). The results were quantities and presented as mean ± SEM of the normalized ROCK levels (n = 3 biological replicates). Statistical significance was determined using a 2-tailed, unpaired Student’s t-test.
Extended Data Figure 9 Direct MYPT1 O-GlcNAcylation is largely responsible for the phenotype.
a, NIH3T3 cells stably expressing either MYPT1 or MYTP1Δ were transfected with RNAi to downregulate endogenous MYTP1. MYTP1-expressing cells were then treated with either DMSO or 5SGlcNAc. Cells under all three sets of conditions were then treated with the indicated concentrations of S1P. The contraction phenotype was visualized using bright-field microscopy. The results were then quantified and are presented as mean ± SEM of the relative culture plate area taken-up by cells in four randomly selected frames. Statistical significance was determined using a 2-way ANOVA test followed by Sidak’s multiple comparisons test. b, NIH3T3 cells stably expressing either MYPT1 or MYTP1Δ were transfected with RNAi to downregulate endogenous MYTP1. MYTP1-expressing cells were then treated with either DMSO or Thiamet-G. Cells under all three sets of conditions were then treated with the indicated concentrations of S1P. The contraction phenotype was visualized using bright-field microscopy. The results were then quantified and are presented as mean ± SEM of the relative culture plate area taken-up by cells in four randomly selected frames. Statistical significance was determined using a 2-way ANOVA test followed by Sidak’s multiple comparisons test.
Extended Data Figure 10 Our experimental model.
a, MYPT1 O-GlcNAcylation inhibits its phosphorylation by ROCK1/2. This maintains MYPT1 phosphatase activity, resulting in inactive MLC and no actin contraction. Loss of O-GlcNAc enables ROCK1/2 to phosphorylate and deactivate MYTP1. b, Therefore, MYPT1 O-GlcNAcylation levels control the sensitivity of cells to the concentration of S1P, where more MYTP1 O-GlcNAcylation requires more S1P to illicit actin contraction and cell detachment.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11.
Supplementary Video 1
O-GlcNAc levels control the contraction of fibroblasts in response to serum.
Supplementary Video 2
O-GlcNAc levels control the contraction of fibroblasts in response to S1P.
Supplementary Data 1
Unprocessed western blots and/or gels for Supplementary Fig. 1.
Supplementary Data 2
Unprocessed western blots and/or gels for Supplementary Fig. 6.
Supplementary Data 3
Unprocessed western blots and/or gels for Supplementary Fig. 10.
Supplementary Data 4
Source data for Supplementary Fig. 2.
Supplementary Data 5
Source data for Supplementary Fig. 3.
Supplementary Data 6
Source data for Supplementary Fig. 7.
Supplementary Data 7
Source data for Supplementary Fig. 8.
Supplementary Data 8
Source data for Supplementary Fig. 10.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Pedowitz, N.J., Batt, A.R., Darabedian, N. et al. MYPT1 O-GlcNAc modification regulates sphingosine-1-phosphate mediated contraction. Nat Chem Biol 17, 169–177 (2021). https://doi.org/10.1038/s41589-020-0640-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0640-8
This article is cited by
-
O-GlcNAcylation determines the translational regulation and phase separation of YTHDF proteins
Nature Cell Biology (2023)