Abstract
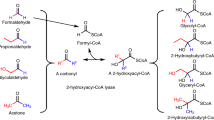
The direct C–H carboxylation of aromatic compounds is an attractive route to the corresponding carboxylic acids, but remains challenging under mild conditions. It has been proposed that the first step in anaerobic microbial degradation of recalcitrant aromatic compounds is a UbiD-mediated carboxylation. In this study, we use the UbiD enzyme ferulic acid decarboxylase (Fdc) in combination with a carboxylic acid reductase to create aromatic degradation-inspired cascade reactions, leading to efficient functionalization of styrene through CO2 fixation. We reveal that rational structure-guided laboratory evolution can expand the substrate scope of Fdc, resulting in activity on a range of mono- and bicyclic aromatic compounds through a single mutation. Selected variants demonstrated 150-fold improvement in the conversion of coumarillic acid to benzofuran + CO2 and unlocked reactivity towards naphthoic acid. Our data demonstrate that UbiD-mediated C–H activation is a versatile tool for the transformation of aryl/alkene compounds and CO2 into commodity chemicals.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Banerjee, A., Dick, G. R., Yoshino, T. & Kanan, M. W. Carbon dioxide utilization via carbonate promoted CH-carboxylation. Nature 531, 215–219 (2016).
Luo, J. & Larrosa, I. C–H carboxylation of aromatic compounds through CO2 fixation. ChemSusChem 10, 3317–3332 (2017).
Hong, J., Li, M., Zhang, J., Sun, B. & Mo, F. C–H bond carboxylation with carbon dioxide. ChemSusChem 12, 6–39 (2019).
Xiao, D. J. et al. A closed cycle for esterifying aromatic hydrocarbons with CO2 and alcohol. Nat. Chem. 11, 940–947 (2019).
Payer, S. E., Faber, K. & Glueck, S. M. Non-oxidative enzymatic (de)carboxylation of (hetero)aromatics and acrylic acid derivatives. Adv. Synth. Cat. 361, 2402–2420 (2019).
Marshall, S. A., Payne, K. A. P. & Leys, D. The UbiX–UbiD system: the biosynthesis and use of prenylated flavin (prFMN). Arch. Biochem. Biophys. 632, 201–221 (2017).
Leys, D. Flavin metamorphosis: cofactor transformation through prenylation. Curr. Opin. Chem. Biol. 47, 117–125 (2018).
White, M. D. et al. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 522, 502–506 (2015).
Marshall, S. A. et al. The UbiX flavin prenyltransferase reaction mechanism resembles class I terpene cyclase chemistry. Nat. Commun. 10, 2357 (2019).
Payne, K. A. P. et al. New cofactor supports α,β-unsaturated acid decarboxylation via 1,3 dipolar cycloaddition. Nature 522, 497–501 (2015).
Bailey, S. S. et al. Enzymatic control of cycloadduct conformation ensures reversible 1,3 dipolar cycloaddition in a prFMN dependent decarboxylase. Nat. Chem. 11, 1049–1057 (2019).
Baunach, M. & Hertweck, C. Natural 1,3-dipolar cycloadditions. Angew. Chem. Int. Ed. 54, 12550–12552 (2015).
Payer, S. E. et al. Regioselective para-carboxylation of catechols with a prenylated flavin dependent decarboxylase. Angew. Chem. Int. Ed. 56, 13893–12897 (2017).
Payne, K. A. P. et al. Enzymatic carboxylation of 2-furoic acid yields 2,5-furandicarboxylic acid (FDCA). ACS Catal. 9, 2854–2865 (2019).
Schuhle, K. & Fuchs, G. Phenylphosphate carboxylase: a new C–C lyase involved in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 186, 4556–4567 (2004).
Meckenstock, R. U. et al. Anaerobic degradation of benzene and polycyclic aromatic hydrocarbons. J. Mol. Microbiol. Biotechnol. 26, 92–118 (2016).
Luo, F. et al. Metatranscriptome of an anaerobic benzene-degrading, nitrate reducing enrichment culture reveals involvement of carboxylation in benzene ring activation. Appl. Environ. Microbiol. 80, 4095–4107 (2014).
Winkler, M. Carboxylic acid reductase enzymes (CARs). Curr. Opin. Chem. Biol. 43, 23–29 (2018).
Finnegan, W. et al. Characterization of carboxylic acid reductases as enzymes in the toolbox for synthetic chemistry. ChemCatChem 9, 1005–1017 (2017).
Wood, A. J. L. et al. Adenylation activity of carboxylic acid reductases enables the synthesis of amides. Angew. Chem. Int. Ed. 56, 14498–14501 (2017).
Zawodny, W. et al. Chemoenzymatic synthesis of substituted azepanes by sequential biocatalytic reduction and organolithium-mediated rearrangement. J. Am. Chem. Soc. 140, 17872–17877 (2018).
Gahloth, D. et al. Structures of carboxylic acid reductase reveal domain dynamics underlying catalysis. Nat. Chem. Biol. 13, 975–981 (2017).
Olah, G. A. Aromatic substitution. XXVIII. Mechanism of electrophilic aromatic substitutions. Acc. Chem. Res. 4, 240–248 (1971).
Murphy, K. E. et al. Precise through-space control of an abiotic electrophilic aromatic substitution reaction. Nat. Commun. 8, 14840 (2017).
Aleku, G. A. et al. A reductive aminase from Aspergillus oryzae. Nat. Chem. 9, 961–969 (2017).
Aleku, G. A., Roberts, G. W. & Leys, D. Biocatalytic reduction of α,β-unsaturated carboxylic acids to allylic alcohols. Green Chem. 22, 3927–3939 (2020).
Winkler, C. K., Faber, K. & Hall, M. Biocatalytic reduction of activated C–C bonds and beyond: emerging trends. Curr. Opin. Chem. Biol. 43, 97–105 (2018).
Villemur, R. Coenzyme A ligases involved in anaerobic biodegradation of aromatic compounds. Can. J. Microbiol. 41, 855–861 (1995).
Wu, S., Zhou, Y. & Li, Z. Biocatalytic selective functionalization of alkenes via single-step and one-pot multi-step reactions. Chem. Commun. 55, 883–896 (2019).
Dabral, S. & Schaub, T. The use of carbon dioxide (CO2) as a building block in organic synthesis from an industrial perspective. Adv. Synth. Catal. 361, 223–246 (2019).
Hepburn, C. et al. The technological and economic prospects for CO2 utilization and removal. Nature 575, 87–97 (2019).
Aleku, G. A. et al. Terminal alkenes from acrylic acid derivatives via non-oxidative enzymatic decarboxylation by ferulic acid decarboxylases. ChemCatChem 10, 3736–3745 (2018).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Grimme, S., Ehrlich, S. & Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 32, 1456–1465 (2011).
Acknowledgements
This work was supported financially by grants BBSRC BB/K017802 (D.L.) and ERC pre-FAB 695013 (D.L.). We acknowledge assistance via use of the Manchester Protein Structure Facility and the Diamond Light Source (proposal nos. MX12788 and MX17773), which contributed to the results presented here. We also acknowledge the assistance given by IT Services and the use of the Computational Shared Facility at The University of Manchester. We thank J. Marshall and N.J. Turner (Manchester) for kindly providing the CfIRED plasmid. D.L. is a Royal Society Wolfson Merit Award holder.
Author information
Authors and Affiliations
Contributions
G.A.A. and R.T.B.-A. performed enzyme inhibition studies, mutagenesis and protein engineering experiments. G.A.A. and I.G. performed crystallization and D.L. determined the associated crystal structures. G.A.A., S.R.D. and D.G. identified suitable carboxylic acid reductases. G.A.A., S.R.D. and G.R.T. identified suitable reductive aminases. G.A.A. and R.T.B.-A. performed decarboxylation biotransformations. G.A.A. constructed CO2 fixation cascades and performed and analyzed cascade biotransformations. A.S. and S.H. performed computational studies. G.A.A. and D.L. designed experiments and wrote the initial draft of the manuscript. D.A.P. advised on all aspects. All authors discussed the results and commented on the manuscript. D.L. initiated and coordinated the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Conversion of styrene to cinnamyl alcohol via carboxylation at near stoichiometric CO2.
a, Reaction scheme for one-pot biocatalytic cascade linking Fdc-catalyzed CO2 fixation to CAR-catalyzed carboxylate reduction at ambient conditions, for the conversion of styrene 1 to cinnamyl alcohol 4via cinnamic acid 2 and cinnamaldehyde 3 intermediates. b, Conversion of styrene to cinnamyl alcohol at various concentration of KHCO3. c, Time-point monitoring of reaction performed at 12.5 mM (2.5 equiv.) KHCO3. For b & c, biotransformation reactions were performed in triplicates, mean conversion values are presented as bars with conversion values from triplicate measurements overlaid as scattered data points. Error bars represent standard deviation from the mean conversion values. d, Comparison of performance of AnFdcubiX used as purified preparation vs as lyophilized cell-free extract (CFE) as the carboxylation catalyst in the one-pot three-step biocatalytic cascade for the conversion of 1 to 4. e, Comparison of AnFdcubiX and ScFdcUbiX as carboxylating catalysts. Similar results were obtained when ScFdcUbiX was used as the carboxylating enzyme under same conditions. [a] Biotransformation conversion values obtained when Fdc was used as cell-free extract (CFE). Reaction conditions, substrate (5 mM), NaPi buffer (50 mM, pH 7.5), 30 °C, 180 r.p.m., 18 h; 2% v/v DMSO. AnFdc enzyme (1 mg mL−1 when used as purified preparation or 10 mg for lyophilized CFE). CARSfp was used as a resting whole-cell preparation to the final OD 600 nm of 30. Conversion values were determined by calibrated HPLC analysis. Trace amount of hydrocinnamylalcohol was also detected. TpCAR = Tsukamurella paurometabola carboxylic acid reductase; AnFdc = ferulic acid decarboxylase from Aspergillus niger; ScFdc = ferulic acid decarboxylase from Saccharomyces cerevisiae.; EcADH: endogenous E. coli alcohol dehydrogenase.
Extended Data Fig. 2 Analysis Fdc-CAR cascade biotransformations for the functionalisation of styrene.
a, AnFdc-TpCAR-EcADH one-pot, three-step cascade: HPLC traces (overlaid) of biotransformation reactions using varying concentration of bicarbonate (10 mM KHCO3- purple, 20 mM - red, 50 mM - blue, 100 mM -green. Retention times: starting material, styrene 1 (13.26 min), cinnamic acid 2 (4.12 min), cinnamaldehyde 3 (6.27 min), cinnamyl alcohol 4 (4.32 min). b, One-pot Fdc-CAR-IRED cascade: GC-MS analysis of biotransformation reactions. Row 1- SrCAR-CfIRED one-pot two-step cascade for the coupling of cinnamic acid 2 with cyclopropylamine to yield N-cinnamylcyclopropanamine 6via carboxylate reduction and reductive amination cascade performed at stoichiometric supply of cofactors. Row 2- SrCAR-CfIRED one-pot two-step cascade for the coupling of cinnamic acid with cyclopropylamine to yield N-cinnamylcyclopropanamine via carboxylate reduction and reductive amination cascade with GDH-based NADPH-recycling system incorporated. Row 3 & 4- AnFdc–SrCAR-CfIRED, one-pot three-step cascade for the conversion of styrene to N-cinnamylcyclopropanamine via carboxylation, carboxylate reduction and reductive amination cascade, and employing GDH-based recycling of NADPH, performed in 100 mM (NH4)HCO3 buffer (Row 3) or 500 mM (NH4)HCO3 buffer (Row 4).
Extended Data Fig. 3 CO2 fixation routes for the conversion of terminal alkenes to amides and amines.
a, CAR-mediated amidation reaction for conversion of cinnamic acid 2 to cinnamide 5. b, One-pot CO2fixation cascade for conversion of styrene 1 to cinnamide 5 using AnFdc WT as the carboxylation catalyst. c, One-pot two-step cascade involving carboxylate reduction and reductive amination for the conversion of carboxylic acid 2 to the corresponding secondary amine 6. Reaction contained cyclopropylamine as the primary amine source. The system incorporated GDH-based recyling of NADPH. d, One-pot three-step CO2 fixation cascade for conversion of styrene 1 to cinnamylcyclopropanamine 6. Reaction contained (NH4)HCO3 as CO2 source and cyclopropylamine as the primary amine source. AnFdc WT was applied as the carboxylation catalyst, SrCAR as the carboxylate reductase, and CfIRED as the reductive aminase. The system incorporated GDH-based recyling of NADPH. SrCAR = Segniliparus rugosus carboxylic acid reductase; AnFdc = ferulic acid decarboxylase from Aspergillus niger; CfIRED = Cystobacter ferrugineus imine reductase; GDH = glucose dehydrogenase.
Extended Data Fig. 4 Screening and identification of competitive inhibitors/alternative substrates of AnFdc.
A library of (hetero)aromatic carboxylic acids screened for inhibition of AnFdc-catalyzed cinnamic acid decarboxylation.
Extended Data Fig. 5 Omit electron density for AnFdc WT and I327S ligand complex structures.
Omit electron density is shown as a cyan mesh contoured at 4 sigma. In case of the complexes with the sulfur containing compound 9, a second mesh (in red) contoured at 12 sigma is shown to illustrate the position of the sulfur atom. In each case the bound cofactor (prFMN or FMN) is shown with yellow carbons, the ligand with green carbon and selected side chains with blue carbons.
Extended Data Fig. 6 Identification of AnFdc variants with improved activity towards (hetero)aromatic carboxylic acids.
a, AnFdc variants displaying improved in vivo decarboxylation activity against representative substrates, identified from HPLC-activity screening of AnFdc M283NDT and I327NDT libraries. b, Substrate profiling study of AnFDC variants I327S/N using purified enzyme preparation of I327S (denoted as A), or I327N (denoted as B) as well as the cell-free extract of I327S (denoted as C). Conversion values were determined by HPLC analysis of biotransformation reactions. Biotransformation reaction conditions were as follows: substrate, 2 mM (or 15 mM for 7), purified enzyme, 1 mg mL−1, DMSO 2-5% v/v, NaPi buffer (50 mM, pH 6, or 100 mM buffer when >10 mM substrate was used), 30 °C, 250 r.p.m., 18 h). [a] Mixture of products, mono-decarboxylated product (40%) and fully decarboxylated product (25%) were detected. c, Specific activity determined spectrophotometrically for AnFdc variants against representative substrates. Activity measurements were performed in triplicate under the same reaction conditions, data are presented as mean specific activity ± standard deviation from mean activity values. d, One-pot artificial CO2 fixation cascade enabling functionalisation of benzofuran 18 to yield benzofuran-2-ylmethanol 20via AnFdc I327S-catalyzed carboxylation. e, One-pot CO2 fixation cascade for conversion of benzofuran 18 to benzofuran-2-carboxamide 21 using AnFdc I327S as the carboxylation catalyst. f, One-pot three-step CO2 fixation cascade for conversion of benzofuran 18 to N-(benzofuran-2-ylmethyl)cyclopropanamine 22. Reaction contained (NH4)HCO3 as CO2 source and cyclopropylamine as the primary amine source. AnFdc I327S was applied as the carboxylation catalyst, TpCAR or SrCAR as the carboxylate reductase, and AspRedAm as the reductive aminase. The system incorporated GDH-based recycling of NADPH. TpCAR = Tsukamurella paurometabola carboxylic acid reductase; SrCAR = Segniliparus rugosus carboxylic acid reductase; AnFdc = ferulic acid decarboxylase from Aspergillus niger; CfIRED = Cystobacter ferrugineus imine reductase; AspRedAm = Aspergillus oryzae reductive aminase; GDH = glucose dehydrogenase; EcADH: endogenous E. coli alcohol dehydrogenase.
Extended Data Fig. 7 GCMS analysis for the decarboxylation of benzothiophene-2-carboxylic acid 9 and 2-naphthoic acid 10 catalyzed by AnFdc I327X.
a, GC-MS chromatogram of control reaction (blue) and AnFdc I327N-catalyzed reaction (black). b, Mass spectra for product detected in the AnFdc I327N-catalyzed reaction, consistent with benzothiophene reference standard. c, AnFdc1 I327N-catalyzed decarboxylation of 2-naphthoic acid 10 to naphthalene and a no enzyme control reaction containing 2-naphthoic acid d, while e, shows the mass spectrum of product peak from biotransformation reaction consistent with the naphthalene reference standard.
Extended Data Fig. 8 One-pot Fdc_CAR_RedAm cascade: GC-MS analysis of biotransformation reactions.
a, SrCAR-CfIRED/AspRedAm one-pot two-step cascade for the coupling of benzofuran-2-carboxylic acid 7 with cyclopropylamine to yield N-(benzofuran-2-ylmethyl)cyclopropanamine via carboxylate reduction and enzymatic reductive amination cascade employing GDH-based recycling of NADPH. b, SrCAR-AspRedAm or SrCAR-CfIRED one-pot two-step cascade for the coupling of benzofuran-2-carboxylic acid 7 with cyclopropylamine to yield N-(benzofuran-2-ylmethyl)cyclopropanamine via carboxylate reduction and reductive amination cascade performed at stoichiometric supply of cofactors; showing reaction intermediates. c, AnFdc I327S-SrCAR-CfIRED, one-pot three-step cascade for the conversion of benzofuran 18 to N-(benzofuran-2-ylmethyl)cyclopropanamine 22via regioselective carboxylation, carboxylate reduction and reductive amination cascade, and employing GDH-based recycling of NADPH, performed in 500 mM (NH4)HCO3.
Extended Data Fig. 9 Computational modeling of the AnFdc I327S decarboxylation of aromatic substrates.
a, Model of the AnFdc I327S active site used for DFT calculations. Red asterisks denote fixed atoms. Overlay of the Int1cyclo (green) and Int1open (blue) models for coumarillic 7. b, and 2-naphthoic acid 10. c, DFT-optimized structures. The Cβ-C4a bond length for the Int1cyclo is 1.66 Å for coumarillic acid and 1.80 Å for 2-naphthoic acid, indicating a high level of strain in the 2-naphthoic acid cycloadduct. The Cβ-C4a bond length for the Int1open is 2.73 Å for coumarillic acid and 2.77 Å for 2-naphthoic acid. d, Natural charge analysis of DFT models of 2-naphthoic acid Sub, Int1open and Int1cyclo. e, Potential energy diagram for the enzyme: substrate to Int2CO2 conversion via Int1 for 2-naphthoic 10 and coumarillic acid 7 determined using DFT cluster models with potential energy normalized to respective enzyme:substrate complexes and approximate transition state energies denoted by double daggers.
Supplementary information
Supplementary Table 1
Data collection and refinement statistics (molecular replacement).
Rights and permissions
About this article
Cite this article
Aleku, G.A., Saaret, A., Bradshaw-Allen, R.T. et al. Enzymatic C–H activation of aromatic compounds through CO2 fixation. Nat Chem Biol 16, 1255–1260 (2020). https://doi.org/10.1038/s41589-020-0603-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0603-0
This article is cited by
-
Light-driven biosynthesis of volatile, unstable and photosensitive chemicals from CO2
Nature Synthesis (2023)
-
Toolbox for the structure-guided evolution of ferulic acid decarboxylase (FDC)
Scientific Reports (2022)
-
Immobilization of carbonic anhydrase for CO2 capture and utilization
Applied Microbiology and Biotechnology (2022)
-
Constructing artificial mimic-enzyme catalysts for carbon dioxide electroreduction
Science China Chemistry (2022)
-
UbiD domain dynamics underpins aromatic decarboxylation
Nature Communications (2021)