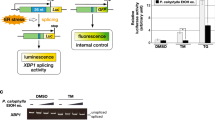
Abstract
Activation of the IRE1/XBP1s signaling arm of the unfolded protein response (UPR) is a promising strategy to correct defects in endoplasmic reticulum (ER) proteostasis implicated in diverse diseases. However, no pharmacologic activators of this pathway identified to date are suitable for ER proteostasis remodeling through selective activation of IRE1/XBP1s signaling. Here, we use high-throughput screening to identify non-toxic compounds that induce ER proteostasis remodeling through IRE1/XBP1s activation. We employ transcriptional profiling to stringently confirm that our prioritized compounds selectively activate IRE1/XBP1s signaling without activating other cellular stress-responsive signaling pathways. Furthermore, we demonstrate that our compounds improve ER proteostasis of destabilized variants of amyloid precursor protein (APP) through an IRE1-dependent mechanism and reduce APP-associated mitochondrial toxicity in cellular models. These results establish highly selective IRE1/XBP1s activating compounds that can be widely employed to define the functional importance of IRE1/XBP1s activity for ER proteostasis regulation in the context of health and disease.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The raw data that support the findings of this study are available within the paper and its Extended Data files (pertaining to Figs. 1, 3 and 4 and Extended Data Figs. 1, 3, 4 and 5), and/or are available from the corresponding author upon reasonable request. The RNA-seq data have been deposited to the public National Center for Biotechnology Information Gene Expression Omnibus repository under the data identifier GSE148802. Source data are provided with this paper.
Code availability
Code for standard open-source DESeq differential gene expression RNA-seq analysis used in R statistical software is available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011).
Bernales, S., Papa, F. R. & Walter, P. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22, 487–508 (2006).
Schroder, M. & Kaufman, R. J. ER stress and the unfolded protein response. Mutat Res. 569, 29–63 (2005).
Patil, C. & Walter, P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349–355 (2001).
Shoulders, M. D. et al. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 3, 1279–1292 (2013).
Lee, A. H., Iwakoshi, N. N. & Glimcher, L. H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459 (2003).
Han, J. et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 (2013).
Chang, T. K. et al. Coordination between two branches of the unfolded protein response determines apoptotic cell fate. Mol. Cell 71, 629–636 e5 (2018).
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008).
Chen, J. J., Genereux, J. C. & Wiseman, R. L. Endoplasmic reticulum quality control and systemic amyloid disease: impacting protein stability from the inside out. IUBMB Life 67, 404–413 (2015).
Wong, M. Y. & Shoulders, M. D. Targeting defective proteostasis in the collagenopathies. Curr. Opin. Chem. Biol. 50, 80–88 (2019).
Zhang, L., Zhang, C. & Wang, A. Divergence and conservation of the major UPR branch IRE1-bZIP signaling pathway across eukaryotes. Sci. Rep. 6, 27362 (2016).
Joshi, A. et al. Molecular mechanisms of human IRE1 activation through dimerization and ligand binding. Oncotarget 6, 13019–13035 (2015).
Korennykh, A. V. et al. The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693 (2009).
Wang, L. et al. Divergent allosteric control of the IRE1α endoribonuclease using kinase inhibitors. Nat. Chem. Biol. 8, 982–989 (2012).
Yoshida, H., Matsui, T., Yamamoto, A., Okada, T. & Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 (2001).
Hollien, J. et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186, 323–331 (2009).
Tam, A. B., Koong, A. C. & Niwa, M. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep. 9, 850–858 (2014).
Bae, D., Moore, K. A., Mella, J. M., Hayashi, S. Y. & Hollien, J. Degradation of Blos1 mRNA by IRE1 repositions lysosomes and protects cells from stress. J. Cell Biol. 218, 1118–1127 (2019).
Valdes, P. et al. Control of dopaminergic neuron survival by the unfolded protein response transcription factor XBP1. Proc. Natl Acad. Sci. USA 111, 6804–6809 (2014).
Zuleta, A., Vidal, R. L., Armentano, D., Parsons, G. & Hetz, C. AAV-mediated delivery of the transcription factor XBP1s into the striatum reduces mutant Huntingtin aggregation in a mouse model of Huntington’s disease. Biochem. Biophys. Res. Commun. 420, 558–563 (2012).
Valenzuela, V. et al. Activation of the unfolded protein response enhances motor recovery after spinal cord injury. Cell Death Dis. 3, e272 (2012).
Cui, H., Deng, M., Zhang, Y., Yin, F. & Liu, J. Geniposide increases unfolded protein response-mediating HRD1 expression to accelerate APP degradation in primary cortical neurons. Neurochem. Res. 43, 669–680 (2018).
Kaneko, M. et al. Loss of HRD1-mediated protein degradation causes amyloid precursor protein accumulation and amyloid-beta generation. J. Neurosci. 30, 3924–3932 (2010).
Chiang, W. C., Messah, C. & Lin, J. H. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol. Biol. Cell 23, 758–770 (2012).
Sifers, R. N. Intracellular processing of alpha1-antitrypsin. Proc. Am. Thorac. Soc. 7, 376–380 (2010).
Ozcan, U. et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306, 457–461 (2004).
Bi, X. et al. Endoplasmic reticulum chaperone GRP78 protects heart from ischemia/reperfusion injury through akt activation. Circ. Res. 122, 1545–1554 (2018).
Mendez, A. S. et al. Endoplasmic reticulum stress-independent activation of unfolded protein response kinases by a small molecule ATP-mimic. eLife 4, e05434 (2015).
Ghosh, R. et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 158, 534–548 (2014).
Plate, L. et al. Small molecule proteostasis regulators that reprogram the ER to reduce extracellular protein aggregation. eLife 5, e15550 (2016).
Iwawaki, T., Akai, R., Kohno, K. & Miura, M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat. Med. 10, 98–102 (2004).
Cross, B. C. et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl Acad. Sci. USA 109, E869–E878 (2012).
Calamini, B. et al. Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol. 8, 185–196 (2011).
Merour, J. Y., Buron, F., Ple, K., Bonnet, P. & Routier, S. The azaindole framework in the design of kinase inhibitors. Molecules 19, 19935–19979 (2014).
Grandjean, J. M. D. et al. Deconvoluting stress-responsive proteostasis signaling pathways for pharmacologic activation using targeted RNA sequencing. ACS Chem. Biol. 14, 784–795 (2019).
Moore, K. & Hollien, J. Ire1-mediated decay in mammalian cells relies on mRNA sequence, structure, and translational status. Mol. Biol. Cell 26, 2873–2884 (2015).
So, J. S. et al. Silencing of lipid metabolism genes through IRE1α-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 16, 487–499 (2012).
Wong, M. Y. et al. XBP1s activation can globally remodel N-glycan structure distribution patterns. Proc. Natl Acad. Sci. USA 115, E10089–E10098 (2018).
Urano, F. et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666 (2000).
Chow, V. W., Mattson, M. P., Wong, P. C. & Gleichmann, M. An overview of APP processing enzymes and products. Neuromolecular Med. 12, 1–12 (2010).
Portelius, E. et al. Mass spectrometric characterization of amyloid-β species in the 7PA2 cell model of Alzheimer’s disease. J. Alzheimers Dis. 33, 85–93 (2013).
Pera, M. et al. Increased localization of APP-C99 in mitochondria-associated ER membranes causes mitochondrial dysfunction in Alzheimer disease. EMBO J. 36, 3356–3371 (2017).
Krako, N. et al. Characterization of mitochondrial dysfunction in the 7PA2 cell model of Alzheimer’s disease. J. Alzheimers Dis. 37, 747–758 (2013).
Rainbolt, T. K., Lebeau, J., Puchades, C. & Wiseman, R. L. Reciprocal degradation of YME1L and OMA1 adapts mitochondrial proteolytic activity during stress. Cell Rep 14, 2041–2049 (2016).
Blackwood, E. A. et al. Pharmacologic ATF6 activation confers global protection in widespread disease models by reprograming cellular proteostasis. Nat. Commun. 10, 187 (2019).
Kroeger, H. et al. The unfolded protein response regulator ATF6 promotes mesodermal differentiation. Sci. Signal 11, eaan5785 (2018).
Casas-Tinto, S. et al. The ER stress factor XBP1s prevents amyloid-beta neurotoxicity. Hum. Mol. Genet. 20, 2144–2160 (2011).
Tufanli, O. et al. Targeting IRE1 with small molecules counteracts progression of atherosclerosis. Proc. Natl Acad. Sci. USA 114, E1395–E1404 (2017).
Rosen, D. A. et al. Modulation of the sigma-1 receptor-IRE1 pathway is beneficial in preclinical models of inflammation and sepsis. Sci. Transl. Med. 11, eaau5266 (2019).
Acknowledgements
We thank J. Lin (UCSD), M.D. Shoulders (MIT), D. Ron (Cambridge) and V. Belyy, W. Li and P. Walter (UCSF) for helpful suggestions and experimental support. We also thank ChemAxon for providing an academic license to perform hierarchical clustering, Library MCS 15.10.12.0 (https://www.chemaxon.com). Funding for this work is provided by the NIH (grant nos. NS095892 and NS092829 to R.L.W.; AG046495 to R.L.W. and J.W.K. and AG063489 to J.M.D.G.) and the Skaggs Institute for Chemical Biology.
Author information
Authors and Affiliations
Contributions
J.M.D.G., A.M., C.B.C., L.P., L.S., T.P.S., J.W.K. and R.L.W. designed the research. J.M.D.G., L.C., B.O.S., A.M., R.J.P., C.B.C. and L.P. performed the chemical, cell biological and biochemical experiments described in this manuscript. E.S. and L.S. performed the HTS to identify IRE1–XBP1s activating compounds. L.S., T.P.S., J.W.K. and R.L.W. provided oversight on the H.T.S. and compound selection. J.M.D.G., E.T.P. and R.L.W. analyzed the RNA-seq data. R.L.W. provided oversight on all aspects of this work. J.M.D.G. and R.L.W. wrote the manuscript with input from other coauthors.
Corresponding author
Ethics declarations
Competing interests
J.W. Kelly is a cofounder of Proteostasis Therapeutics Inc. A patent has been submitted for compounds identified through work described in this manuscript. This patent includes R.L. Wiseman, J.W. Kelly, J.M.D. Grandjean, L. Plate and C.B. Cooley as coinventors. No other author has competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 HTS to identify preferential IRE1–XBP1s activating compounds.
a, Schematic of the XBP1-Renilla luciferase (XBP1-RLuc) splicing reporter used in our high-throughput screen to identify small molecule activators of IRE1/XBP1s signaling. b, Graph showing XBP1-RLuc splicing reporter activation in HEK293TREX cells incubated with Tg (500 nM) in the presence or absence of 4µ8c (64 µM) for 16 hrs. Error bars show SD for n = 3 replicates. c, Plot showing XBP1-RLuc activation in HEK293TREX cells stably expressing the XBP1-RLuc reporter treated with the 10,114 small molecules (6 µM; 18 hrs) identified as hits in the primary screen. Luminescence is shown as % signal relative to treatment with Tg (500 nM; 18 hrs). d, Venn diagram of overlap of compounds identified to activate the IRE1-dependent XBP1-RLuc splicing reporter, the ATF6-selective ERSE-FLuc reporter, or the HSF1-dependent HSP70-FLuc reporter via high-throughput screening. e, Plot showing XBP1-RLuc activation in HEK293TREX cells (% signal compared to that observed with 500 nM Tg, 18 hrs) versus promiscuity score for our top 638 compounds identified by HTS. The promiscuity score reports on the number of assays performed at the TSRI-FL Screening Center where each compound was identified as a positive hit. f, Plot showing IC50 for toxicity as measured by CellTiterGlo luminescence calculated from titration screening of 638 hit compounds. The dashed red lines indicates IC50 = 3 µM. g, Plot showing IRE1-dependent XBP1-RLuc activation versus ATF6-selective ERSE-Fluc activation in HEK293TREX cells (% signal compared to that observed with 500 nM Tg, 18 hrs) for the subset of our top 638 compounds that were also identified to activate the ATF6-selective ERSE-FLuc reporter. The dashed red line indicates equal XBP1s-Rluc and ERSE-Fluc activation. h, Plot showing XBP1-RLuc activation in HEK293TREX cells (% signal compared to that observed with 500 nM Tg, 18 hrs) versus EC50 of XBP1-RLuc activation in HEK293TREX cells calculated from titration screening of hit 638 compounds. The dashed red lines indicates 20% Rluc activation and Rluc EC50 = 3 µM.
Extended Data Fig. 2 Compound-dependent IRE1–XBP1s activation requires IRE1 autophosphorylation.
a, Plot showing XBP1-RLuc activation in HEK293TREX cells (% signal compared to that observed with 500 nM Tg, 18 hrs) treated for 18 hrs with the indicated concentrations of prioritized IRE1/XBP1s activator. Error bars represent SD for n = 3 replicates. b, Graph showing qPCR of the XBP1s target gene DNAJB9 in HEK293T cells treated for 4 hrs with Tg (500 nM) in the presence or absence of 4µ8c (64 µM). Error bars show SE for n = 3. P-values were calculated using one-tailed Student’s t-test. **p < 0.01. c, Graph showing qPCR of the ATF6 target gene BiP in HEK293T cells treated for 4 hrs with Tg (500 nM) in the presence or absence of 4µ8c (64 µM). Error bars show SE for n = 3. P-values were calculated using one-tailed Student’s t-test. d, Graph showing qPCR of the PERK target gene CHOP in HEK293T cells treated for 4 hrs with Tg (500 nM) in the presence or absence of 4µ8c (64 µM). Error bars show SE for n = 3. P-values were calculated using one-tailed Student’s t-test. e, Graph showing qPCR of the PERK target gene CHOP in HEK293T cells treated for 4 hrs with prioritized IRE1/XBP1s activators (10 µM) or Tg (500 nM), in the presence or absence of 4µ8c (32 µM). Error bars show 95% CI for n = 3 replicates. f, Graph showing qPCR analysis of the ATF6 target gene BiP in HEK293T cells treated for 4 hrs with indicated compound (10 µM), or Tg (500 nM), in the presence or absence of 4µ8c (32 µM). Error bars show 95% CI for n = 3 replicates. g, cDNA gel showing splicing of XBP1 mRNA in WT MEF or Ire1-/- MEF cells treated with IXA1, IXA4, or IXA6 (10 µM) for 4 hrs.
Extended Data Fig. 3 Transcriptional profiling of compounds IXA1, IXA4 and IXA6 shows preferential induction of IRE1–XBP1s target genes.
a, Graph showing log2 Fold change mRNA levels from RNA-seq for the RIDD target SCARA3 in HEK293T cells treated with 10 µM IXA1, IXA4, IXA6, or 500 nM Tg for 4 hrs. Error bars represent SD for n = 3 replicates. b, Graph showing log2 Fold change mRNA levels from RNA-seq for the RIDD target BLOC1S1 in HEK293T cells treated with 10 µM IXA1, IXA4, IXA6, or 500 nM Tg for 4 hrs. Error bars represent SD for n = 3 replicates. c, Graph showing log2 Fold change mRNA levels from RNA-seq for the RIDD target COL6A1 in HEK293T cells treated with 10 µM IXA1, IXA4, IXA6, or 500 nM Tg for 4 hrs. Error bars represent SD for n = 3 replicates.
Extended Data Fig. 4 Compounds IXA4 and IXA6 show selectivity for IRE1–XBP1s-dependent ER proteostasis remodeling (part 1).
a, Gene Ontology (GO) analysis of differentially expressed genes from RNA-seq for HEK293T cells treated with IXA1 (10 µM, 4hrs). Top 15 entries with lowest FDR are shown. See Supplementary Table 4 for full GO analysis. b, Graph showing fold change mRNA levels from RNA-seq of target genes activated downstream of the UPR, HSR, OSR, and other stress signaling pathways in HEK293T cells treated with IXA1 (10 µM) for 4 hrs. The composition of these gene sets is shown in Source Data Fig. 3. P-values were calculated using one-way ANOVA compared to ‘Other’. ****p < 0.0001. c, Graph showing fold change mRNA levels from RNA-seq of target genes activated downstream of the UPR, HSR, OSR, and other stress signaling pathways in HEK293T cells treated with IXA4 (10 µM) for 4 hrs. The composition of these genesets is shown in Source Data Fig. 3. P-values were calculated using one-way ANOVA compared to ‘Other’. ****p < 0.0001. d, Graph showing fold change mRNA levels from RNA-seq of target genes activated downstream of the UPR, HSR, OSR, and other stress signaling pathways in HEK293T cells treated with IXA6 (10 µM) for 4 hrs. The composition of these genesets is shown in Source Data Fig. 3. P-values were calculated using one-way ANOVA compared to ‘Other’. ****p < 0.0001. e, Bar graph showing fold change mRNA levels of the IRE1/XBP1s targets DNAJB9, SEC24D, and HSPA13 from RNA-seq of HEK293DAX cells expressing dox-inducible XBP1s treated with dox (1 µg/mL) for 4 hr or HEK293T cells treated with compounds IXA1, IXA4, or IXA6 (10 µM) for 4 hrs. f, Venn diagram of genes upregulated >1.2 fold (adjusted p-value <0.05) in HEK293T cells treated with compound IXA1 (10 µM) for 4 hrs in comparison to genes induced >1.2 fold (adjusted p-value < 0.05) in HEK293DAX cells treated with doxycycline (1 µg/mL) for 4 hrs. Genes listed in purple are top overlapping targets between conditions. g, Graph showing log2 Fold Change mRNA levels from RNA-seq of the 10 non-overlapping genes activated in cells treated with IXA4 (10 µM) compared to dox-inducible XBP1s. Log2 Fold change mRNA levels of these genes in cells treated with IXA6 (10 µM) are also included. h, Graph showing qPCR of the LRRCC1 gene in HEK293T cells treated with IXA4 (10 µM) in the presence or absence of 4µ8c (64 µM) for 4 hrs. Error bars show SE for n = 3. i, Graph showing qPCR of the CETN3 gene in HEK293T cells treated with IXA4 (10 µM) in the presence or absence of 4µ8c (64 µM) for 4 hrs. Error bars show SE for n = 3.
Extended Data Fig. 5 Compounds IXA4 and IXA6 show selectivity for IRE1–XBP1s-dependent ER proteostasis remodeling (part 2).
a, Heat map of top 100 genes upregulated in HEK293T cells treated for 4 hrs with IXA4 (10 µM) compared to dox-inducible XBP1s or TMP-stabilized DHFR.ATF6 in HEK293DAX cells treated with doxycycline (1 µg/mL) or TMP (10 µM) for 4 hr. b, Heat map of top 100 genes upregulated in HEK293T cells treated for 4 hrs with IXA6 (10 µM) compared to dox-inducible XBP1s or TMP-stabilized DHFR.ATF6 in HEK293DAX cells treated with doxycycline (1 µg/mL) or TMP (10 µM) for 4 hrs. c, Heat map of top 100 genes upregulated in HEK293T cells treated for 4 hr with IXA1 (10 µM) compared to dox-inducible XBP1s or TMP-stabilized DHFR.ATF6 in HEK293DAX cells treated with doxycycline (1 µg/mL) or TMP (10 µM) for 4 hrs.
Extended Data Fig. 6 Compounds IXA4 and IXA6 show selectivity for IRE1–XBP1s-dependent ER proteostasis remodeling (part 3).
a, Representative immunoblots of XBP1s, PERK, eIF2α, and BiP in lysates prepared from HEK293T cells treated with vehicle, thapsigargin (1 µM), IXA4 (10 µM), or IXA6 (10 µM) for 4 hrs. b, Graph showing qPCR of the XBP1 target gene DNAJB9, ATF6 target gene BiP, and PERK target gene CHOP in Huh-7 cells treated with IXA4 (10 µM) or IXA6 (10 µM) in the presence or absence of 4µ8c (64 µM) for 4 hrs. Error bars show SE for n = 3 replicates. Statistics calculated from one-tailed Student’s t-test. *p < 0.05, **p < 0.01. c, Graph showing qPCR of the XBP1s target gene DNAJB9, ATF6 target gene BiP, and PERK target gene CHOP in SHSY5Y cells treated with IXA4 (10 µM) or IXA6 (10 µM) in the presence or absence of 4µ8c (64 µM) for 4 hrs. Error bars show SE for n = 4 replicates. Statistics calculated from one-tailed Student’s t-test. *p < 0.05, **p < 0.01. d, Graph showing relative signal of phosphorylated target proteins by phosphokinase array dot immunoblotting in HEK293T cells treated with IXA4 (10 µM) for 4 hrs. Error bars show SD for n = 2.
Extended Data Fig. 7 Compound IXA4 increases degradation of APP mutants (part 1).
a, Representative immunoblots and quantification of SEC24D in lysates prepared from HEK293T cells treated with vehicle, IXA1, IXA4, or IXA6 (10 µM, 18 hrs). Error bars show SD for n = 3 replicates. P-values were calculated using one-tailed Student’s t-test. *p < 0.05, **p < 0.01. b, Plot of log2 Fold Change mRNA levels from RNA-seq in cells treated with IXA6 (10 µM, 4 hrs) of proteostasis factors found in the ER, cytosol/nucleus, or mitochondria. The composition of these proteostasis genesets is shown in Source Data Fig. 3. c, Graph showing the Fold change mRNA ratio of Xbp1s:Xbp1u by qPCR in CHO7PA2 cells treated with IXA4 (10 µM) or thapsigargin (1 µM) in the presence or absence of 4µ8c (64 µM) for 4 hrs. Error bars show SE for n = 3 replicates. Statistics calculated from one-tailed Student’s t-test. *p < 0.05, **p < 0.01. d, Graph showing relative CellTiterGlo luminescence from CHO7PA2 cells treated with IRE1/XBP1s activators IXA4, or IXA6 (10 µM) in the presence or absence of 4µ8c (32 µM). Cells were treated for 18 hrs, media was then replaced and conditioned in the presence of treatments for 24 hrs before measuring ATP levels. Luminescence signal was normalized to that observed in untreated controls. Error bars represent SE for n = 3 replicates. e, Graph showing cell counts (106cells/mL) of CHO7PA2 cells treated with IXA4 or IXA6 (10 µM) in the presence or absence of 4µ8c (64 µM) for 24 hrs. f, Immunoblot of PARP in lysates collected from CHO7PA2 cells treated with IXA4 or IXA6 (10 µM) or staurosporine (STS) (1 µM) for 24 hrs. g, Graph showing relative signal from ELISA of secreted Aβ peptide from conditioned media prepared on CHO7WD10 cells treated with IXA4 or IXA6 (10 µM). Cells were pretreated for 18 hrs with compounds. Media was then replaced and conditioned in the presence of compounds for 24 hrs before harvesting the media for ELISA. Secreted Aβ was normalized to that observed in untreated controls. Error bars represent SE for n = 3 replicates. h, Graph showing relative CellTiterGlo luminescence from CHO7WD10 cells treated with IRE1/XBP1s activators IXA4 or IXA6 (10 µM). Cells were treated for 18 hrs, media was then replaced and conditioned in the presence of treatments for 24 hrs before measuring ATP levels. Luminescence signal was normalized to that observed in untreated controls. Error bars represent SE for n = 3 replicates.
Extended Data Fig. 8 Compound IXA4 increases degradation of APP mutants (part 2).
a, Representative immunoblot of mutant APP in media and lysates collected from CHO7PA2 cells treated with IXA1 or IXA4 (10 µM) in the presence or absence of 4µ8c (64 µM). Cells were treated for 18 hrs, media was then replaced and conditioned in the presence of treatments for 24 hrs before harvesting. b, Representative immunoblot of mutant APP in media and lysates collected from CHO7PA2 cells treated with IXA4 or IXA6 (10 µM) in the presence or absence of MG132 (10 µM) for 18hrs. c, Quantification of immunoblots represented in panel b of relative APP signal in lysates or conditioned media from CHO7PA2 cells treated with IRE1/XBP1s activators IXA4 or IXA6 (10 µM) in the presence or absence of MG132 (10 µM) for 18hrs. Error bars represent SE for n = 4 replicates. Statistics calculated from one-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. d, Representative autoradiogram showing the [35S] metabolic labeling of mutant APP in CHO7PA2 cells treated with IXA6 (10 µM) for 16 hrs prior to 30 min labeling. Media and lysates were collected at 0, 1, or 2 hrs and [35S]-labeled mutant APP was isolated by immunopurification. The experimental protocol is shown above. Fraction remaining was calculated as described in Fig. 5d and fraction secretion was calculated as in Fig. 5e. Error bars represent SD for n = 3 replicates. P-values were calculated from one-tailed Student’s t-test. *p < 0.05.
Extended Data Fig. 9 The IRE1–XBP1s activator IXA4 rescues mitochondrial defects in SHSY5Y cells expressing disease-relevant APP mutants.
a, Graph showing qPCR of the XBP1s target gene DNAJB9 in SHSY5Y cells transiently expressing empty vector (Mock) or Swedish mutant APP (APPSW) in the presence or absence of IXA4 (10 µM) for 72 hrs. Error bars show SE for n = 2. Statistics calculated from one-tailed Student’s t-test. *p < 0.05, **p < 0.01. b, Graph showing qPCR of the PERK target gene CHOP in SHSY5Y cells transiently expressing empty vector (Mock) or APPSW in the presence or absence of IXA4 (10 µM) for 72 hrs. Error bars show SE for n = 2. c, Representative histograms showing TMRE staining of SHSY5Y cells transiently expressing empty vector (Mock) or wild-type APP (APPWT) in the presence or absence of IXA4 (10 µM) for 72 hrs.
Supplementary information
Supplementary Information
Supplementary Fig. 1, Table 1 and descriptions of Tables 2–4.
Supplementary Table 2
Representative 128 compounds from structural analysis of small molecule IRE1/XBP1s activators.
Supplementary Table 3
Internal library numbers for selected IRE1/XBP1s activators.
Supplementary Table 4
GO analysis of prioritized IRE1/XBP1s activator transcriptional profiles.
Source data
Source Data Fig. 1
Small molecules HTS data for 638 hits.
Source Data Fig. 2
Unprocessed western blot and DNA gel for C&D.
Source Data Fig. 3
Whole transcriptome RNA-seq aligned counts and lists of proteostasis gene sets.
Source Data Fig. 5
Unprocessed S35 autoradiogram.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Unprocessed western blots for a and f.
Source Data Extended Data Fig. 8
Unprocessed western blots and S35 autoradiogram for a, b and d.
Rights and permissions
About this article
Cite this article
Grandjean, J.M.D., Madhavan, A., Cech, L. et al. Pharmacologic IRE1/XBP1s activation confers targeted ER proteostasis reprogramming. Nat Chem Biol 16, 1052–1061 (2020). https://doi.org/10.1038/s41589-020-0584-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0584-z
This article is cited by
-
Imbalanced unfolded protein response signaling contributes to 1-deoxysphingolipid retinal toxicity
Nature Communications (2023)
-
Control of immune cell function by the unfolded protein response
Nature Reviews Immunology (2023)
-
SEC-seq: association of molecular signatures with antibody secretion in thousands of single human plasma cells
Nature Communications (2023)
-
Hypoxia Affects Autophagy in Human Umbilical Vein Endothelial Cells via the IRE1 Unfolded Protein Response
Current Medical Science (2023)
-
Dual RNase activity of IRE1 as a target for anticancer therapies
Journal of Cell Communication and Signaling (2023)