Abstract
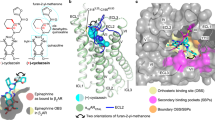
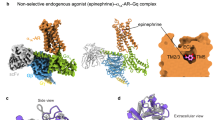
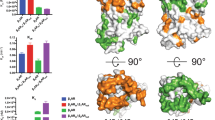
Most drugs acting on G-protein-coupled receptors target the orthosteric binding pocket where the native hormone or neurotransmitter binds. There is much interest in finding allosteric ligands for these targets because they modulate physiologic signaling and promise to be more selective than orthosteric ligands. Here we describe a newly developed allosteric modulator of the β2-adrenergic receptor (β2AR), AS408, that binds to the membrane-facing surface of transmembrane segments 3 and 5, as revealed by X-ray crystallography. AS408 disrupts a water-mediated polar network involving E1223.41 and the backbone carbonyls of V2065.45 and S2075.46. The AS408 binding site is adjacent to a previously identified molecular switch for β2AR activation formed by I3.40, P5.50 and F6.44. The structure reveals how AS408 stabilizes the inactive conformation of this switch, thereby acting as a negative allosteric modulator for agonists and positive allosteric modulator for inverse agonists.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Atomic coordinates and structure factors have been deposited in the PDB under accession code 6OBA.
References
Kruse, A. C. et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106 (2013).
Digby, G. J., Shirey, J. K. & Conn, P. J. Allosteric activators of muscarinic receptors as novel approaches for treatment of CNS disorders. Mol. Biosyst. 6, 1345–1354 (2010).
Mohr, K., Trankle, C. & Holzgrabe, U. Structure/activity relationships of M2 muscarinic allosteric modulators. Recept. Channels 9, 229–240 (2003).
Wootten, D., Christopoulos, A. & Sexton, P. M. Emerging paradigms in GPCR allostery: implications for drug discovery. Nat. Rev. Drug Discov. 12, 630–644 (2013).
Thal, D. M., Glukhova, A., Sexton, P. M. & Christopoulos, A. Structural insights into G-protein-coupled receptor allostery. Nature 559, 45–53 (2018).
Ahn, S. et al. Allosteric ‘beta-blocker’ isolated from a DNA-encoded small molecule library. Proc. Natl Acad. Sci. USA 114, 1708–1713 (2017).
Ahn, S. et al. Small-molecule positive allosteric modulators of the beta2-adrenoceptor isolated from DNA-encoded libraries. Mol. Pharmacol. 94, 850–861 (2018).
Liu, X. et al. Mechanism of intracellular allosteric beta2AR antagonist revealed by X-ray crystal structure. Nature 548, 480–484 (2017).
Liu, X. et al. Mechanism of beta2AR regulation by an intracellular positive allosteric modulator. Science 364, 1283–1287 (2019).
Rasmussen, S. G. et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469, 175–180 (2011).
Masureel, M. et al. Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist. Nat. Chem. Biol. 14, 1059–1066 (2018).
Ballesteros, J. A. & Weinstein, H. in Methods in Neurosciences Vol. 25 (ed. Sealfon, S. C.) 366–428 (Academic Press, 1995).
Leach, K., Sexton, P. M. & Christopoulos, A. Allosteric GPCR modulators: taking advantage of permissive receptor pharmacology. Trends Pharmacol. Sci. 28, 382–389 (2007).
Aurelio, L. et al. Allosteric modulators of the adenosine A1 receptor: synthesis and pharmacological evaluation of 4-substituted 2-amino-3-benzoylthiophenes. J. Med. Chem. 52, 4543–4547 (2009).
Christopoulos, A. & Kenakin, T. G protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 54, 323–374 (2002).
Srivastava, A. et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature 513, 124–127 (2014).
Robertson, N. et al. Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature 553, 111–114 (2018).
Liu, H. et al. Orthosteric and allosteric action of the C5a receptor antagonists. Nat. Struct. Mol. Biol. 25, 472–481 (2018).
Roth, C. B., Hanson, M. A. & Stevens, R. C. Stabilization of the human beta2-adrenergic receptor TM4-TM3-TM5 helix interface by mutagenesis of Glu122(3.41), a critical residue in GPCR structure. J. Mol. Biol. 376, 1305–1319 (2008).
Schonegge, A. M. et al. Evolutionary action and structural basis of the allosteric switch controlling beta2AR functional selectivity. Nat. Commun. 8, 2169(2017).
Du, Y. et al. Assembly of a GPCR-G protein complex. Cell 177, e1211 (2019).
Szlenk, C. T., Gc, J. B. & Natesan, S. Does the lipid bilayer orchestrate access and binding of ligands to transmembrane orthosteric/allosteric sites of G protein-coupled receptors? Mol. Pharmacol. 96, 527–541 (2019).
Lu, S. & Zhang, J. Small molecule allosteric modulators of G-protein-coupled receptors: drug-target interactions. J. Med. Chem. 62, 24–45 (2019).
Pettersen, E. F. et al. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Klicić, J. J., Friesner, R. A., Liu, S.-Y. & Guida, W. C. Accurate prediction of acidity constants in aqueous solution via density functional theory and self-consistent reaction field methods. J. Phys. Chem. A. 106, 1327–1335 (2002).
Bochevarov, A. D., Watson, M. A., Greenwood, J. R. & Philipp, D. M. Multiconformation, density functional theory-based pKa prediction in application to large, flexible organic molecules with diverse functional groups. J. Chem. Theory Comput. 12, 6001–6019 (2016).
Yu, H. S., Watson, M. A. & Bochevarov, A. D. Weighted averaging scheme and local atomic descriptor for pKa prediction based on density functional theory. J. Chem. Inf. Model 58, 271–286 (2018).
Lomize, M. A., Lomize, A. L., Pogozheva, I. D. & Mosberg, H. I. OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 (2006).
Wolf, M. G., Hoefling, M., Aponte-Santamaria, C., Grubmuller, H. & Groenhof, G. g_membed: efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J. Comput. Chem. 31, 2169–2174 (2010).
Case, D. A. et al. AMBER 2017 (University of California, San Francisco, 2017).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Dickson, C. J. et al. Lipid14: the Amber lipid force field. J. Chem. Theory Comput. 10, 865–879 (2014).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Berendsen, H. J. C., Grigera, J. R. & Straatsma, T. P. The missing term in effective pair potentials. J. Phys. Chem. 91, 6269–6271 (1987).
Frisch, M. J. et al. Gaussian 09, revision B.01 (Gaussian, Inc., Wallingford, CT, 2009).
Bayly, C. I., Cieplak, P., Cornell, W. D. & Kollman, P. A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the Resp model. J. Phys. Chem. 97, 10269–10280 (1993).
Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald—an n.log(n) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Schrödinger Release 2018-1: Maestro, Schrödinger, LLC, New York, NY, 2018.
Rosenbaum, D. M. et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science 318, 1266–1273 (2007).
Kobilka, B. K. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal. Biochem. 231, 269–271(1995).
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protocol. 4, 706–731 (2009).
Kabsch, W. XDS. Acta Crystallogr. Sect. D 66, 125–132 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D 66, 213–221 (2010).
Ten Eyck, L. F. Fast Fourier transform calculation of electron density maps. Methods Enzymol. 115, 324–337 (1985).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D 67, 235–242 (2011).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D 66, 12–21 (2010).
Hubner, H. et al. Structure-guided development of heterodimer-selective GPCR ligands. Nat. Commun. 7, 12298 (2016).
DeVree, B. T. et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature 535, 182–186 (2016).
Whorton, M. R. et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proceed. Natl Acad. Sci. USA 104, 7682–7687 (2007).
Vedel, L., Brauner-Osborne, H. & Mathiesen, J. M. A cAMP biosensor-based high-throughput screening assay for identification of Gs-coupled GPCR ligands and phosphodiesterase inhibitors. J. Biomol. Screen 20, 849–857 (2015).
Acknowledgements
We acknowledge support from the US NIH grant nos. GM106990 (B.K.S., P.G. and R.K.S.), and the DFG Grants Gm 13/10 and GRK 1910 (P.G.), and Beijing Advanced Innovation Center for Structural Biology, Tsinghua University (X.L.), as well as the compute resources provided by the RRZE. B.K.K. is a Chan Zuckerberg Biohub Investigator. We thank D. Steffen and M. Gilardi for help with confocal microscopy. We also thank A. Christopoulos for his critical insight with the analysis of cooperativity.
Author information
Authors and Affiliations
Contributions
X.L. expressed and purified the receptor, crystallized the receptor–ligand complex and solved the crystal structure. K.H. performed the automatic data collection and processing. A.S., D.D. and M.S. synthesized and analytically characterized the chemical compounds. H.H., M.J.C., R.A.M., J.M., X.L., D.D. and X.X. performed ligand binding and signaling experiments. M.K. performed docking studies. J.K. performed MD simulations. The manuscript was written by B.K.K., X.L. and P.G. with editing and suggestions from R.K.S. and input from J.K. and H.H., P.G. supervised chemical synthesis of compounds, B.K.K., R.K.S. and P.G. supervised binding and signaling experiments. B.K.S. supervised docking. X.L. supervised structure determination. The project was conceived by B.K.K., P.G., R.K.S. and B.K.S.
Corresponding authors
Ethics declarations
Competing interests
B.K.K. is a cofounder of and consultant for ConfometRx, Inc.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–5, Figs. 1–10 and Note
Rights and permissions
About this article
Cite this article
Liu, X., Kaindl, J., Korczynska, M. et al. An allosteric modulator binds to a conformational hub in the β2 adrenergic receptor. Nat Chem Biol 16, 749–755 (2020). https://doi.org/10.1038/s41589-020-0549-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0549-2
This article is cited by
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
Structural insights into the activation and inhibition of CXC chemokine receptor 3
Nature Structural & Molecular Biology (2024)
-
Cryo-electron microscopy for GPCR research and drug discovery in endocrinology and metabolism
Nature Reviews Endocrinology (2024)
-
GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway
Nature Structural & Molecular Biology (2024)
-
Constrained catecholamines gain β2AR selectivity through allosteric effects on pocket dynamics
Nature Communications (2023)