Abstract
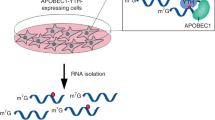
The inert chemical property of RNA modification N6-methyladenosine (m6A) makes it very challenging to detect. Most m6A sequencing methods rely on m6A-antibody immunoprecipitation and cannot distinguish m6A and N6,2′-O-dimethyladenosine modification at the cap +1 position (cap m6Am). Although the two antibody-free methods (m6A-REF-seq/MAZTER-seq and DART-seq) have been developed recently, they are dependent on m6A sequence or cellular transfection. Here, we present an antibody-free, FTO-assisted chemical labeling method termed m6A-SEAL for specific m6A detection. We applied m6A-SEAL to profile m6A landscapes in humans and plants, which displayed the known m6A distribution features in transcriptome. By doing a comparison with all available m6A sequencing methods and specific m6A sites validation by SELECT, we demonstrated that m6A-SEAL has good sensitivity, specificity and reliability for transcriptome-wide detection of m6A. Given its tagging ability and FTO’s oxidation property, m6A-SEAL enables many applications such as enrichment, imaging and sequencing to drive future functional studies of m6A and other modifications.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data have been deposited in GSE129979.
Code availability
Custom Bash, Perl and R codes used for data analysis are available at https://github.com/WYeast.
References
Yue, Y., Liu, J. & He, C. RNA N 6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355 (2015).
Jia, G. et al. N 6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N 6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Ping, X.-L. et al. Mammalian WTAP is a regulatory subunit of the RNA N 6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Wang, X. et al. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Wang, X. et al. N 6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Li, A. et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444 (2017).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Hsu, P. J. et al. Ythdc2 is an N 6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127 (2017).
Nachtergaele, S. & He, C. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 52, 349–372 (2018).
Li, Z. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N 6-methyladenosine RNA demethylase. Cancer Cell 31, 127–141 (2017).
Lichinchi, G. et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 20, 666–673 (2016).
Huang, Y. et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell 35, 677–691 (2019).
Tirumuru, N. et al. N 6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 5, e15528 (2016).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Bastian, L. et al. Single-nucleotide resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Chen, K. et al. High-resolution N 6-methyladenosine m6A map using photo-crosslinking-assisted m6A sequencing. Angew. Chem. Int. Ed. 127, 1607–1610 (2015).
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015).
Molinie, B. et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 13, 692 (2016).
Zhang, Z. et al. Single-base mapping of m6A by an antibody-independent method. Sci. Adv. 5, eaax0250 (2019).
Garcia-Campos, M. A. et al. Deciphering the ‘m6A Code’ via antibody-independent quantitative profiling. Cell 178, 731–747.e16 (2019).
Meyer, K. D. DART-seq: an antibody-free method for global m6A detection. Nat. Methods 16, 1275–1280 (2019).
Fu, Y. et al. FTO-mediated formation of N 6-hydroxymethyladenosine and N 6-formyladenosine in mammalian RNA. Nat. Commun. 4, 1798 (2013).
Lu, K. et al. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 132, 3388–3399 (2010).
You, X. J. et al. Determination of RNA hydroxylmethylation in mammals by mass spectrometry analysis. Anal. Chem. 91, 10477–10483 (2019).
Huber, S. M. et al. Formation and abundance of 5-hydroxymethylcytosine in RNA. Chem. Bio. Chem. 16, 752–755 (2015).
Liu, N. et al. Probing N 6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Liu, J. et al. N 6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 31, 580–586 (2020).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2016).
Zhang, F. et al. The subunit of RNA N 6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 15, e1008120 (2019).
Li, Y. et al. Transcriptome-wide N 6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 11, 1180–1188 (2014).
Wei, L.-H. et al. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30, 968–985 (2018).
Xiao, Y. et al. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N 6-methyladenosine modification. Angew. Chem. Int. Ed. 57, 15995–16000 (2018).
Sun, H., Zhang, M., Li, K., Bai, D. & Yi, C. Cap-specific, terminal N 6-methylation by a mammalian m6Am methyltransferase. Cell Res. 29, 80–82 (2019).
Sendinc, E. et al. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell 75, 620–630.e9 (2019).
Boulias, K. et al. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol. Cell 75, 631–643.e8 (2019).
Fu, Y. et al. N 6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161, 879–892 (2015).
Greer, E. L. et al. DNA methylation on N 6-Adenine in C. elegans. Cell 161, 868–878 (2015).
Zhang, G. Q. et al. N 6-methyladenine DNA modification in Drosophila. Cell 161, 893–906 (2015).
Wu, T. P. et al. DNA methylation on N 6-adenine in mammalian embryonic stem cells. Nature 532, 329–333 (2016).
Xiao, C. L. et al. N 6-methyladenine DNA modification in the human genome. Mol. Cell 71, 306–318.e7 (2018).
Jia, G. et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 582, 3313–3319 (2008).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Bailey, T. L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–1659 (2011).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Cui, X. et al. Guitar: an R/bioconductor package for gene annotation guided transcriptomic analysis of RNA-related genomic features. BioMed. Res. Int. 2016, 8367534 (2016).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Acknowledgements
We acknowledge X. Lei for discussing the thiol-addition reaction mechanism and S. Liu, J. Meng and H. Liu for helping with data analysis. This work was supported by the National Basic Research Program of China (grant nos. 2019YFA0802201 and 2017YFA0505201), the National Natural Science Foundation of China (nos. 21822702, 21820102008 and 21432002) and the State Key Laboratory of Drug Research.
Author information
Authors and Affiliations
Contributions
G.J. and Y.W. conceived the project and designed the experiments. Y.W. performed the experiments and data analysis with the help of Y.X., S.D. and Q.Y. G.J. and Y.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
A patent application has been filed by Peking University for the technology disclosed in this publication.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplemental Information
Supplementary Table 1–3, Figs. 1–25 and Notes 1–3.
Supplementary Dataset 1
Contains two worksheets, followed by explanations for each column.
Supplementary Dataset 2
Contains one worksheet, followed by an explanation for each column.
Rights and permissions
About this article
Cite this article
Wang, Y., Xiao, Y., Dong, S. et al. Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat Chem Biol 16, 896–903 (2020). https://doi.org/10.1038/s41589-020-0525-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0525-x
This article is cited by
-
Decoding epitranscriptomic regulation of viral infection: mapping of RNA N6-methyladenosine by advanced sequencing technologies
Cellular & Molecular Biology Letters (2024)
-
RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy
Signal Transduction and Targeted Therapy (2024)
-
GLORI for absolute quantification of transcriptome-wide m6A at single-base resolution
Nature Protocols (2024)
-
Single-cell m6A mapping in vivo using picoMeRIP–seq
Nature Biotechnology (2024)
-
Recent advances in the plant epitranscriptome
Genome Biology (2023)