Abstract
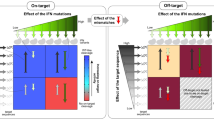
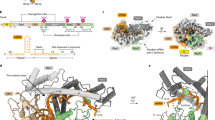
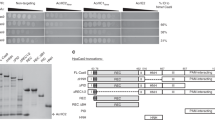
Anti-CRISPR (Acr) proteins are powerful tools to control CRISPR–Cas technologies. However, the available Acr repertoire is limited to naturally occurring variants. Here, we applied structure-based design on AcrIIC1, a broad-spectrum CRISPR–Cas9 inhibitor, to improve its efficacy on different targets. We first show that inserting exogenous protein domains into a selected AcrIIC1 surface site dramatically enhances inhibition of Neisseria meningitidis (Nme)Cas9. Then, applying structure-guided design to the Cas9-binding surface, we converted AcrIIC1 into AcrIIC1X, a potent inhibitor of the Staphylococcus aureus (Sau)Cas9, an orthologue widely applied for in vivo genome editing. Finally, to demonstrate the utility of AcrIIC1X for genome engineering applications, we implemented a hepatocyte-specific SauCas9 ON-switch by placing AcrIIC1X expression under regulation of microRNA-122. Our work introduces designer Acrs as important biotechnological tools and provides an innovative strategy to safeguard CRISPR technologies.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Vectors encoding AcrIIC1X, AcrIIC1X* as well as the AcrIIC1-mCherry chimera will be made available via Addgene (plasmids nos. 128112–128114). Annotated vector sequences (GenBank files) are provided in the Supplementary Data 1. Additional data can be obtained from the corresponding authors on reasonable request.
Code availability
Code and data for the computational domain assembly and the design of the improved AcrIIC1 point mutants will be made available on GitHub (https://github.com/LPDI-EPFL/AcrIIC1X).
References
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Bondy-Denomy, J., Pawluk, A., Maxwell, K. L. & Davidson, A. R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013).
Bondy-Denomy, J. et al. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 (2015).
Pawluk, A., Bondy-Denomy, J., Cheung, V. H., Maxwell, K. L. & Davidson, A. R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio 5, e00896 (2014).
Basgall, E. M. et al. Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology 164, 464–474 (2018).
Bubeck, F. et al. Engineered anti-CRISPR proteins for optogenetic control of CRISPR–Cas9. Nat. Methods 15, 924–927 (2018).
Hoffmann, M. D. et al. Cell-specific CRISPR–Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic Acids Res. 47, e75 (2019).
Nakamura, M. et al. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat. Commun. 10, 194 (2019).
Shin, J. et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 3, e1701620 (2017).
Aschenbrenner, S. et al. Coupling Cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity. Sci. Adv. 6, eaay0187 (2019).
Rauch, B. J. et al. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 168, 150–158.e10 (2016).
Pawluk, A. et al. Inactivation of CRISPR–Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol 1, 16085 (2016).
Pawluk, A. et al. Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829–1838 e9 (2016).
Lee, J. et al. Potent Cas9 inhibition in bacterial and human cells by AcrIIC4 and AcrIIC5 anti-CRISPR proteins. mBio 9, e02321–18 (2018).
Hynes, A. P. et al. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun. 9, 2919 (2018).
Watters, K. E., Fellmann, C., Bai, H. B., Ren, S. M. & Doudna, J. A. Systematic discovery of natural CRISPR–Cas12a inhibitors. Science 362, 236–239 (2018).
Marino, N. D. et al. Discovery of widespread type I and type V CRISPR–Cas inhibitors. Science 362, 240–242 (2018).
Garcia, B. et al. Anti-CRISPR AcrIIA5 potently inhibits all Cas9 homologs used for genome editing. Cell Rep. 29, 1739–1746 e5 (2019).
Harrington, L. B. et al. A broad-spectrum inhibitor of CRISPR-Cas9. Cell 170, 1224–1233.e15 (2017).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168–e168 (2014).
Sentmanat, M. F., Peters, S. T., Florian, C. P., Connelly, J. P. & Pruett-Miller, S. M. A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 8, 888 (2018).
Edraki, A. et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell 73, 714–726 e4 (2019).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Senís, E. et al. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 9, 1402–1412 (2014).
Schmidt, F. & Grimm, D. CRISPR genome engineering and viral gene delivery: a case of mutual attraction. Biotechnol. J. 10, 258–272 (2015).
Leaver-Fay, A. et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 (2011).
Lee, J. et al. Tissue-restricted genome editing in vivo specified by microRNA-repressible anti-CRISPR proteins. RNA 25, 1421–1431 (2019).
Hirosawa, M., Fujita, Y. & Saito, H. Cell-type-specific CRISPR activation with microRNA-responsive AcrllA4 switch. ACS Synth. Biol. 8, 1575–1582 (2019).
Osuna, B. A. et al. Listeria phages induce Cas9 degradation to protect lysogenic genomes. Preprint at bioRxiv https://doi.org/10.1101/787200 (2019).
Sesterhenn, F. et al. De novo protein design enables precise induction of functional antibodies in vivo. Preprint at bioRxiv https://doi.org/10.1101/685867 (2019).
Song, G. et al. AcrIIA5 inhibits a broad range of Cas9 orthologs by preventing DNA target cleavage. Cell Rep. 29, 2579–2589 e4 (2019).
Agudelo, D. et al. Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. Genome Res. 30, 107–117 (2018).
Watters, K.E. et al. Potent CRISPR-Cas9 inhibitors from Staphylococcus genomes. Preprint at bioRxiv https://doi.org/10.1101/799403 (2019).
Huang, P. S. et al. RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS ONE 6, e24109 (2011).
Canutescu, A. A. & Dunbrack, R. L. Jr. Cyclic coordinate descent: a robotics algorithm for protein loop closure. Protein Sci. 12, 963–972 (2003).
Coutsias, E. A., Seok, C., Jacobson, M. P. & Dill, K. A. A kinematic view of loop closure. J. Comput. Chem. 25, 510–528 (2004).
Mandell, D. J., Coutsias, E. A. & Kortemme, T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 6, 551–552 (2009).
Bonet, J., Harteveld, Z., Sesterhenn, F., Scheck, A. & Correia, B. E. rstoolbox—a Python library for large-scale analysis of computational protein design data and structural bioinformatics. BMC Bioinf. 20, 240 (2019).
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005).
Fleishman, S. J. et al. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS ONE 6, e20161 (2011).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Engler, C. & Marillonnet, S. Combinatorial DNA assembly using Golden Gate cloning. Methods Mol. Biol. 1073, 141–156 (2013).
Amrani, N. et al. NmeCas9 is an intrinsically high-fidelity genome-editing platform. Genome Biol. 19, 214 (2018).
Herrmann, A. K. et al. A robust and all-inclusive pipeline for shuffling of adeno-associated viruses. ACS Synth. Biol. 8, 194–206 (2019).
Börner, K. et al. Robust RNAi enhancement via human Argonaute-2 overexpression from plasmids, viral vectors and cell lines. Nucleic Acids Res. 41, e199 (2013).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 18, 529 (2017).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
We thank the Synthetic Biology (IPMB, Heidelberg University), Virus-Host Interactions (Heidelberg University Clinics) and the Protein Design and Immunoengineering groups (EPFL, Lausanne) for helpful discussions; K. Börner (Heidelberg University Clinics, Heidelberg, Germany) and K. Rippe (DKFZ, Heidelberg, Germany) for providing cell lines; and K. Niopek for critical reading of the manuscript. We thank the EPFL’s Scientific IT and Application Support Center for their support on the computational infrastructure. We thank the Protein Production and Structure Core facility at EPFL for their support on the protein biophysical characterization experiments. This study was funded by the Helmholtz association, the German Research Foundation (DFG) and the Federal Ministry of Education and Research (BMBF) (R.E.). D.G. is grateful for funding from the German Center for Infection Research (DZIF, grant no. TTU-HIV 04.803 and grant no. TTU-HIV 04.815) and the Cystic Fibrosis Foundation Therapeutics (CFFT, grant no. GRIMM15XX0). C. Schmelas, D.G. and R.E. acknowledge funding from the Transregional Collaborative Research Center TRR179 (DFG, Projektnummer 272983813). C. Schmelas and D.G. acknowledge additional funding by the Cluster of Excellence CellNetworks (DFG, grant no. EXC81). B.E.C. is a grantee from the European Research Council (starting grant no. 716058), the Swiss National Science Foundation and the Biltema Foundation. Part of the computational simulations were performed at the CSCS Swiss National Supercomputing Centre through a grant obtained by B.E.C. Z.H. is supported by a grant from the Swiss National Science Foundation. Z.H. and S.R. are supported by a grant from the National Center of Competence in Research in Chemical Biology. Y.W. is supported by the Natural Science Foundation of China (grant nos. 31930065, 31725008, 31630015, 31571335 and 31700662).
Author information
Authors and Affiliations
Contributions
D.N. conceived the initial idea and refined it together with J.U.z.B. and B.E.C.. J.M., C. Schmelas, C. Stengl, S.A. and D.N. designed and performed experiments. S.A. and M.D.H. performed AAV production. S.R. and S.G. purified the Acrs and performed protein-biochemical characterization. Z.H., J.U.z.B., A.S. and B.E.C. performed in silico structural analysis and modeling. W.S. and Y.W. designed and performed in vitro DNA binding and cleavage assays. D.G. provided expertise on AAVs and SauCas9. R.E. supported the computational and experimental work. B.E.C. and D.N. jointly directed the work. B.E.C. and D.N. wrote the manuscript with support from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–18 and Tables 1–3.
Supplementary Data 1
Annotated vector sequences (GenBank files).
Rights and permissions
About this article
Cite this article
Mathony, J., Harteveld, Z., Schmelas, C. et al. Computational design of anti-CRISPR proteins with improved inhibition potency. Nat Chem Biol 16, 725–730 (2020). https://doi.org/10.1038/s41589-020-0518-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0518-9
This article is cited by
-
Characterization of the AcrIIC1 anti‒CRISPR protein for Cas9‒based genome engineering in E. coli
Communications Biology (2023)
-
A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1
Nature Communications (2022)
-
Inhibition of base editors with anti-deaminases derived from viruses
Nature Communications (2022)
-
Base editing: advances and therapeutic opportunities
Nature Reviews Drug Discovery (2020)