Abstract
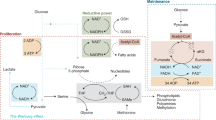
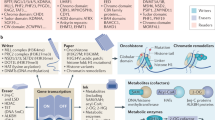
In eukaryotes, chromatin remodeling and post-translational modifications (PTMs) shape the local chromatin landscape to establish permissive and repressive regions within the genome, orchestrating transcription, replication, and DNA repair in concert with other epigenetic mechanisms. Though cellular nutrient signaling encompasses a huge number of pathways, recent attention has turned to the hypothesis that the metabolic state of the cell is communicated to the genome through the type and concentration of metabolites in the nucleus that are cofactors for chromatin-modifying enzymes. Importantly, both epigenetic and metabolic dysregulation are hallmarks of a range of diseases, and this metabolism–chromatin axis may yield a well of new therapeutic targets. In this Perspective, we highlight emerging themes in the inter-regulation of the genome and metabolism via chromatin, including nonenzymatic histone modifications arising from chemically reactive metabolites, the expansion of PTM diversity from cofactor-promiscuous chromatin-modifying enzymes, and evidence for the existence and importance of subnucleocytoplasmic metabolite pools.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kornberg, R. D. Structure of chromatin. Annu. Rev. Biochem. 46, 931–954 (1977).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001).
Allis, C.D., Caparros, M.L., Jenuwein, T. & Reinberg, D. Epigenetics (Cold Spring Harbor Laboratory Press, 2015).
Zoghbi, H. Y. & Beaudet, A. L. Epigenetics and human disease. Cold Spring Harb. Perspect. Biol. 8, a019497 (2016).
Chi, P., Allis, C. D. & Wang, G. G. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010).
Cai, L., Sutter, B. M., Li, B. & Tu, B. P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 (2011).
Shyh-Chang, N. et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226 (2013).
Ryu, K. W. et al. Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 360, eaan5780 (2018). This paper details the subcellular compartmentalization of NAD + synthesis and how these nuclear and cytoplasmic NAD + pools regulate adipogenic transcriptional programs.
Li, S. et al. Serine and SAM responsive complex SESAME regulates histone modification crosstalk by sensing cellular metabolism. Mol. Cell 60, 408–421 (2015).
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Yang, M., Soga, T. & Pollard, P. J. Oncometabolites: linking altered metabolism with cancer. J. Clin. Invest. 123, 3652–3658 (2013).
Ye, C., Sutter, B. M., Wang, Y., Kuang, Z. & Tu, B. P. A metabolic function for phospholipid and histone methylation. Mol. Cell 66, 180–193.e8 (2017).
Bulusu, V. et al. Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Reports 18, 647–658 (2017).
Schvartzman, J. M., Thompson, C. B. & Finley, L. W. S. Metabolic regulation of chromatin modifications and gene expression. J. Cell Biol. 217, 2247–2259 (2018).
Suganuma, T. & Workman, J. L. Chromatin and metabolism. Annu. Rev. Biochem. 87, 27–49 (2018).
Li, X., Egervari, G., Wang, Y., Berger, S. L. & Lu, Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat. Rev. Mol. Cell Biol. 19, 563–578 (2018).
Galligan, J. J. & Marnett, L. J. Histone adduction and its functional impact on epigenetics. Chem. Res. Toxicol. 30, 376–387 (2017).
Kulkarni, R. A. et al. Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem. Biol. 24, 231–242 (2017).
Simithy, J. et al. Characterization of histone acylations links chromatin modifications with metabolism. Nat. Commun. 8, 1141–1153 (2017).
Galligan, J. J. et al. Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. J. Am. Chem. Soc. 136, 11864–11866 (2014). This paper identifies 4-oxo-2-nonenal adducts on histones both in vitro and in cells and shows that treatment of macrophages with lipopolysaccharide enhances this modification on H3K27.
Jiang, T., Zhou, X., Taghizadeh, K., Dong, M. & Dedon, P. C. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc. Natl. Acad. Sci. USA 104, 60–65 (2007).
Guedes, S., Vitorino, R., Domingues, M. R. M., Amado, F. & Domingues, P. Glycation and oxidation of histones H2B and H1: in vitro study and characterization by mass spectrometry. Anal. Bioanal. Chem. 399, 3529–3539 (2011).
Galligan, J. J. et al. Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc. Natl Acad. Sci. USA 115, 9228–9233 (2018). This paper and ref. 53 characterize glycation as an abundant histone PTM modulated by glycolytic activity and regulated by the enzymes GLO-1 and DJ-1.
Carrier, E. J., Zagol-Ikapitte, I., Amarnath, V., Boutaud, O. & Oates, J. A. Levuglandin forms adducts with histone h4 in a cyclooxygenase-2-dependent manner, altering its interaction with DNA. Biochemistry 53, 2436–2441 (2014). This paper identifies adducts of levuglandin, a byproduct of fatty acid peroxidation, on histones as a function of arachidonic acid treatment and COX-2 expression and describes how these modifications affect nucleosome structure.
Long, E. K., Olson, D. M. & Bernlohr, D. A. High-fat diet induces changes in adipose tissue trans-4-oxo-2-nonenal and trans-4-hydroxy-2-nonenal levels in a depot-specific manner. Free Radic. Biol. Med. 63, 390–398 (2013).
Preidis, G. A. et al. The undernourished neonatal mouse metabolome reveals evidence of liver and biliary dysfunction, inflammation, and oxidative stress. J. Nutr. 144, 273–281 (2014).
Allaman, I., Bélanger, M. & Magistretti, P. J. Methylglyoxal, the dark side of glycolysis. Front. Neurosci. 9, 23–34 (2015).
Ajith, T. A. & Vinodkumar, P. Advanced glycation end products: association with the pathogenesis of diseases and the current therapeutic advances. Curr. Clin. Pharmacol. 11, 118–127 (2016).
Singh, R., Barden, A., Mori, T. & Beilin, L. Advanced glycation end-products: a review. Diabetologia 44, 129–146 (2001).
Palsson-McDermott, E. M. & O’Neill, L. A. J. The Warburg effect then and now: from cancer to inflammatory diseases. BioEssays 35, 965–973 (2013).
Bonadonna, R. C. Alterations of glucose metabolism in type 2 diabetes mellitus. An overview. Rev. Endocr. Metab. Disord. 5, 89–97 (2004).
Maze, I. et al. Critical role of histone turnover in neuronal transcription and Plasticity. Neuron 87, 77–94 (2015).
Wagner, G. R. & Payne, R. M. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288, 29036–29045 (2013).
Baeza, J., Smallegan, M. J. & Denu, J. M. Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem. Biol. 10, 122–128 (2015).
Xie, Z. et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107 (2012).
Tan, M. et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617 (2014).
Kaczmarska, Z. et al. Structure of p300 in complex with acyl-CoA variants. Nat. Chem. Biol. 13, 21–29 (2017).
Bao, X. et al. Glutarylation of histone H4 lysine 91 regulates chromatin Dynamics. Mol. Cell 76, 660–675.e9 (2019).
Wang, Y. et al. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017). This paper reports KAT2A (also called GCN5) as a histone succinyltransferase that binds to the α-KGDH enzyme in the nucleus to utilize the locally produced succinyl-CoA to deposit the mark on H3K79 at gene promoters.
Ishiguro, T. et al. Malonylation of histone H2A at lysine 119 inhibits Bub1-dependent H2A phosphorylation and chromosomal localization of shugoshin proteins. Sci. Rep. 8, 7671 (2018).
Röhrig, F. & Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 (2016).
Zhu, X. & Sayre, L. M. Long-lived 4-oxo-2-enal-derived apparent lysine michael adducts are actually the isomeric 4-ketoamides. Chem. Res. Toxicol. 20, 165–170 (2007).
Jin, J., He, B., Zhang, X., Lin, H. & Wang, Y. SIRT2 reverses 4-oxononanoyl lysine modification on histones. J. Am. Chem. Soc. 138, 12304–12307 (2016).
Mont, S. et al. Accumulation of isolevuglandin-modified protein in normal and fibrotic lung. Sci. Rep. 6, 24919 (2016).
Boutaud, O., Andreasson, K. I., Zagol-Ikapitte, I. & Oates, J. A. Cyclooxygenase-dependent lipid-modification of brain proteins. Brain Pathol. 15, 139–142 (2005).
Schröter, D. & Höhn, A. Role of advanced glycation end products in carcinogenesis and their therapeutic implications. Curr. Pharm. Des. 24, 5245–5251 (2018).
Lu, C. et al. Advanced glycation endproduct (AGE) receptor 1 is a negative regulator of the inflammatory response to AGE in mesangial cells. Proc. Natl. Acad. Sci. USA 101, 11767–11772 (2004).
Talasz, H., Wasserer, S. & Puschendorf, B. Nonenzymatic glycation of histones in vitro and in vivo. J. Cell. Biochem. 85, 24–34 (2002).
Gugliucci, A. & Bendayan, M. Histones from diabetic rats contain increased levels of advanced glycation end products. Biochem. Biophys. Res. Commun. 212, 56–62 (1995).
Ashraf, J. M. et al. 3-Deoxyglucosone: a potential glycating agent accountable for structural alteration in H3 histone protein through generation of different AGEs. PLoS One 10, e0116804 (2015).
Ashraf, J. M. et al. Physicochemical analysis of structural alteration and advanced glycation end products generation during glycation of H2A histone by 3-deoxyglucosone. IUBMB Life 66, 686–693 (2014).
Zheng, Q. et al. Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat. Commun. 10, 1289 (2019). This paper and ref. 24 characterize glycation as an abundant histone PTM modulated by glycolytic activity and regulated by the enzymes GLO-1 and DJ-1.
Richarme, G. et al. Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science 357, 208–211 (2017).
Cao, J., Lou, S., Ying, M. & Yang, B. DJ-1 as a human oncogene and potential therapeutic target. Biochem. Pharmacol. 93, 241–250 (2015).
Qi, W. et al. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat. Med. 23, 753–762 (2017).
Sabari, B. R., Zhang, D., Allis, C. D. & Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 18, 90–101 (2017).
Farrelly, L. A. et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567, 535–539 (2019). This paper reports serotonylation of H3Q5 and details its impact on transcription in serotonergic neurons through this PTM’s co-occurrence with H3K4me3.
Chen, Y. et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 (2007).
Huang, H. et al. p300-mediated lysine 2-hydroxyisobutyrylation regulates Glycolysis. Mol. Cell 70, 663–678.e6 (2018).
Sabari, B. R. et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 (2015). This paper characterizes histone crotonylation as a function of the ratio of acetyl-CoA:crotonyl-CoA showing that this modification is more abundant under conditions of low acetyl-CoA and modulates gene expression in a manner distinct from histone acetylation.
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019). This paper characterizes lactylation of histone lysines as an epigenetic modification that directly stimulates gene transcription and is regulated by lactate production.
Ringel, A. E. & Wolberger, C. Structural basis for acyl-group discrimination by human Gcn5L2. Acta Crystallogr. D Struct. Biol. 72, 841–848 (2016).
Lee, J. V. et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 20, 306–319 (2014).
Xie, Z. et al. Metabolic Regulation of Gene Expression by Histone Lysine β-Hydroxybutyrylation. Mol. Cell 62, 194–206 (2016). This paper reports lysine β-hydroxybutyrylation as a new histone modification that is enriched at active gene promoters and is upregulated under starvation conditions in mice that produce high levels of the ketone body β-hydroxybutyrate.
Barnes, C. E., English, D. M. & Cowley, S. M. Acetylation & Co: an expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem. 63, 97–107 (2019).
Zhao, S., Zhang, X. & Li, H. Beyond histone acetylation-writing and erasing histone acylations. Curr. Opin. Struct. Biol. 53, 169–177 (2018).
Goudarzi, A. et al. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol. Cell 62, 169–180 (2016).
Kebede, A. F. et al. Histone propionylation is a mark of active chromatin. Nat. Struct. Mol. Biol. 24, 1048–1056 (2017). This paper characterizes histone propionylation and butyrylation, finding the H3K14pr and H3K14bu marks enriched at active gene promoters. Whereas H3K14bu was unaffected by butyryl-CoA levels, H3K14pr levels were modulated at a function of cellular propionyl-CoA.
Nelson, D.L. & Cox, M.M. Lehninger Principles of Biochemistry. (W. H. Freeman, 2017).
Stilling, R. M. et al. The neuropharmacology of butyrate: the bread and butter of the microbiota-gut-brain axis? Neurochem. Int. 99, 110–132 (2016).
Liu, X. et al. High-resolution metabolomics with acyl-CoA profiling reveals widespread remodeling in response to diet. Mol. Cell. Proteomics 14, 1489–1500 (2015).
Smestad, J., Erber, L., Chen, Y. & Maher, L. J. Chromatin succinylation correlates with active gene expression and is perturbed by defective TCA cycle metabolism. iScience 2, 63–75 (2018).
Wang, Y., Guo, Y. R., Xing, D., Tao, Y. J. & Lu, Z. Supramolecular assembly of KAT2A with succinyl-CoA for histone succinylation. Cell Discov. 4, 47 (2018).
Dai, L. et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 10, 365–370 (2014).
Huang, H. et al. Landscape of the regulatory elements for lysine 2-hydroxyisobutyrylation pathway. Cell Res. 28, 111–125 (2018).
Calvani, R. et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int. J. Obes.(Lond) 34, 1095–1098 (2010).
Diaz, S. O. et al. Metabolic biomarkers of prenatal disorders: an exploratory NMR metabonomics study of second trimester maternal urine and blood plasma. J. Proteome Res. 10, 3732–3742 (2011).
Li, M. et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 105, 2117–2122 (2008).
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Li, Y. et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell 62, 181–193 (2016).
Liu, S. et al. Chromodomain protein CDYL acts as a crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis. Mol. Cell 67, 853–866.e5 (2017).
Hummerich, R., Thumfart, J.-O., Findeisen, P., Bartsch, D. & Schloss, P. Transglutaminase-mediated transamidation of serotonin, dopamine and noradrenaline to fibronectin: evidence for a general mechanism of monoaminylation. FEBS Lett. 586, 3421–3428 (2012).
Kim, J.-H. et al. Histone cross-linking by transglutaminase. Biochem. Biophys. Res. Commun. 293, 1453–1457 (2002).
Rubí, B. & Maechler, P. Minireview: new roles for peripheral dopamine on metabolic control and tumor growth: let’s seek the balance. Endocrinology 151, 5570–5581 (2010).
Maintz, L. & Novak, N. Histamine and histamine intolerance. Am. J. Clin. Nutr. 85, 1185–1196 (2007).
Sudo, N. Biogenic amines: signals between commensal microbiota and gut physiology. Front. Endocrinol. (Lausanne) 10, 504 (2019).
Barros, L. F. & Martínez, C. An enquiry into metabolite domains. Biophys. J. 92, 3878–3884 (2007).
Zecchin, A., Stapor, P. C., Goveia, J. & Carmeliet, P. Metabolic pathway compartmentalization: an underappreciated opportunity? Curr. Opin. Biotechnol. 34, 73–81 (2015).
Kabachinski, G. & Schwartz, T. U. The nuclear pore complex—structure and function at a glance. J. Cell Sci. 128, 423–429 (2015).
Saks, V., Beraud, N. & Wallimann, T. Metabolic compartmentation – a system level property of muscle cells: real problems of diffusion in living cells. Int. J. Mol. Sci. 9, 751–767 (2008).
Kekenes-Huskey, P. M., Scott, C. E. & Atalay, S. Quantifying the influence of the crowded cytoplasm on small molecule diffusion. J. Phys. Chem. B 120, 8696–8706 (2016).
Fulton, A. B. How crowded is the cytoplasm? Cell 30, 345–347 (1982).
Sivanand, S., Viney, I. & Wellen, K. E. Spatiotemporal control of acetyl-CoA metabolism in chromatin regulation. Trends Biochem. Sci. 43, 61–74 (2018).
Nagaraj, R. et al. Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell 168, 210–223.e11 (2017).
Sivanand, S. et al. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol. Cell 67, 252–265.e6 (2017).
Matsuda, S. et al. Nuclear pyruvate kinase M2 complex serves as a transcriptional coactivator of arylhydrocarbon receptor. Nucleic Acids Res. 44, 636–647 (2016).
Greenwald, E. C., Mehta, S. & Zhang, J. Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 118, 11707–11794 (2018).
Chantranupong, L., Wolfson, R. L. & Sabatini, D. M. Nutrient-sensing mechanisms across evolution. Cell 161, 67–83 (2015).
Evans, R. M. & Mangelsdorf, D. J. Nuclear receptors, RXR, and the Big Bang. Cell 157, 255–266 (2014).
Acknowledgements
We thank current and former members of the Muir laboratory for discussions and comments. Some of the work discussed herein was performed in the author’s laboratory and was supported by National Institutes of Health (NIH) Grants R37 GM086868, R01 GM107047 and P01 CA196539. K.L.D. was supported by an NIH Research Service Awards (5F32CA206418).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Diehl, K.L., Muir, T.W. Chromatin as a key consumer in the metabolite economy. Nat Chem Biol 16, 620–629 (2020). https://doi.org/10.1038/s41589-020-0517-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0517-x
This article is cited by
-
Acylspermidines are conserved mitochondrial sirtuin-dependent metabolites
Nature Chemical Biology (2024)
-
Metabolic regulation of epigenetic drug resistance
Nature Chemical Biology (2023)
-
Lactate as a major epigenetic carbon source for histone acetylation via nuclear LDH metabolism
Experimental & Molecular Medicine (2023)
-
Histone acetyltransferase NAA40 modulates acetyl-CoA levels and lipid synthesis
BMC Biology (2022)
-
Modulation of cellular processes by histone and non-histone protein acetylation
Nature Reviews Molecular Cell Biology (2022)