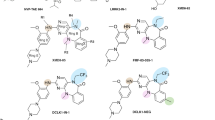
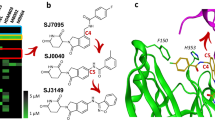
Abstract
Doublecortin like kinase 1 (DCLK1) is an understudied kinase that is upregulated in a wide range of cancers, including pancreatic ductal adenocarcinoma (PDAC). However, little is known about its potential as a therapeutic target. We used chemoproteomic profiling and structure-based design to develop a selective, in vivo-compatible chemical probe of the DCLK1 kinase domain, DCLK1-IN-1. We demonstrate activity of DCLK1-IN-1 against clinically relevant patient-derived PDAC organoid models and use a combination of RNA-sequencing, proteomics and phosphoproteomics analysis to reveal that DCLK1 inhibition modulates proteins and pathways associated with cell motility in this context. DCLK1-IN-1 will serve as a versatile tool to investigate DCLK1 biology and establish its role in cancer.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
KINOMEscan and KiNativ data are provided in Supplementary Datasets 1 and 2. Cell line RNA-sequencing data has been deposited to the NCBI GEO (accession number GSE140490). Cell line and deidentified patient-derived organoid RNA-sequencing analyzed data files are provided in Supplementary Dataset 3. Cell line and deidentified patient-derived organoid mass spectrometry-based proteomics and phosphoproteomics analyzed data files are provided in Supplementary Datasets 4 and 5.
References
Fedorov, O., Muller, S. & Knapp, S. The (un)targeted cancer kinome. Nat. Chem. Biol. 6, 166–169 (2010).
Westphalen, C. B., Quante, M. & Wang, T. C. Functional implication of Dclk1 and Dclk1-expressing cells in cancer. Small GTPases 8, 164–171 (2017).
Nishio, K. et al. Doublecortin and CaM kinase-like-1 as an independent prognostic factor in patients with resected pancreatic carcinoma. World J. Gastroenterol. 23, 5764–5772 (2017).
Adamska, A., Domenichini, A. & Falasca, M. Pancreatic ductal adenocarcinoma: current and evolving therapies. Int. J. Mol. Sci. 18, E1338 (2017).
Westphalen, C. B. et al. Dclk1 defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell Stem Cell 18, 441–455 (2016).
Brubaker, D. K. et al. Proteogenomic network analysis of context-specific KRAS signaling in mouse-to-human cross-species translation. Cell Syst. 9, 258–270.e6 (2019).
Antolin, A. A. et al. Objective, quantitative, data-driven assessment of chemical probes. Cell Chem. Biol. 25, 194–205.e5 (2018).
Deng, X. et al. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat. Chem. Biol. 7, 203–205 (2011).
Yang, Q. et al. Pharmacological inhibition of BMK1 suppresses tumor growth through PML. Cancer Cell 18, 258–267 (2010).
Deng, X. et al. Discovery of a benzo(e)pyrimido-(5,4-b)(1,4)diazepin-6(11H)-one as a potent and selective inhibitor of Big MAP Kinase 1. ACS Med. Chem. Lett. 2, 195–200 (2011).
Lin, E. C. et al. ERK5 kinase activity is dispensable for cellular immune response and proliferation. Proc. Natl Acad. Sci. USA 113, 11865–11870 (2016).
Wang, J. et al. Structural and atropisomeric factors governing the selectivity of pyrimido-benzodiazipinones as inhibitors of kinases and bromodomains. ACS Chem. Biol. 13, 2438–2448 (2018).
Weygant, N. et al. Small molecule kinase inhibitor LRRK2-IN-1 demonstrates potent activity against colorectal and pancreatic cancer through inhibition of doublecortin-like kinase 1. Mol. Cancer 13, 103 (2014).
Xu, Y. & Vakoc, C. R. Targeting cancer cells with BET bromodomain inhibitors. Cold Spring Harb. Perspect. Med. 7, a026674 (2017).
Hoang, V. T. et al. Oncogenic signaling of MEK5-ERK5. Cancer Lett. 392, 51–59 (2017).
Lin, A. et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 11, eaaw8412 (2019).
Miduturu, C. V. et al. High-throughput kinase profiling: a more efficient approach toward the discovery of new kinase inhibitors. Chem. Biol. 18, 868–879 (2011).
Patricelli, M. P. et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem. Biol. 18, 699–710 (2011).
Patel, O. et al. Biochemical and structural insights into doublecortin-like kinase domain 1. Structure 24, 1550–1561 (2016).
Davis, M. I. et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 (2011).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Vasta, J. D. et al. Quantitative, wide-spectrum kinase profiling in live cells for assessing the effect of cellular ATP on target engagement. Cell Chem. Biol. 25, 206–214.e11 (2018).
Beno, B. R., Yeung, K. S., Bartberger, M. D., Pennington, L. D. & Meanwell, N. A. A survey of the role of noncovalent sulfur interactions in drug design. J. Med. Chem. 58, 4383–4438 (2015).
Chen, H. et al. Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site. Acta Crystallogr. D: Biol. Crystallogr. 72, 682–693 (2016).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Nabet, B. et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 (2018).
Janes, M. R. et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018).
Muzumdar, M. D. et al. Survival of pancreatic cancer cells lacking KRAS function. Nat. Commun. 8, 1090 (2017).
Gilmartin, A. G. et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 17, 989–1000 (2011).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
McDonald, E. R.3rd et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577–592.e10 (2017).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576.e16 (2017).
Gendoo, D. M. A. et al. Whole genomes define concordance of matched primary, xenograft, and organoid models of pancreas cancer. PLoS Comput. Biol. 15, e1006596 (2019).
Tiriac, H. et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 8, 1112–1129 (2018).
Foster, S. A. et al. Activation mechanism of oncogenic deletion mutations in BRAF, EGFR, and HER2. Cancer Cell 29, 477–493 (2016).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545 (2005).
Drost, J. & Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 18, 407–418 (2018).
Ting, H. A. & von Moltke, J. The immune function of tuft cells at gut mucosal surfaces and beyond. J. Immunol. 202, 1321–1329 (2019).
Keller, S. et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 (2012).
Erazo, T. et al. Canonical and kinase activity-independent mechanisms for extracellular signal-regulated kinase 5 (ERK5) nuclear translocation require dissociation of Hsp90 from the ERK5-Cdc37 complex. Mol. Cell. Biol. 33, 1671–1686 (2013).
Wang, B. et al. ATXN1L, CIC, and ETS transcription factors modulate sensitivity to MAPK pathway inhibition. Cell Rep. 18, 1543–1557 (2017).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Banker, G. & Goslin, K. Culturing Nerve Cells. (The MIT Press: 1998).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Loven, J. et al. Revisiting global gene expression analysis. Cell 151, 476–482 (2012).
Paulo, J. A. Nicotine alters the proteome of two human pancreatic duct cell lines. JOP 15, 465–474 (2014).
Paulo, J. A. & Gygi, S. P. Nicotine-induced protein expression profiling reveals mutually altered proteins across four human cell lines. Proteomics 17, 1–2 (2017).
Navarrete-Perea, J., Yu, Q., Gygi, S. P. & Paulo, J. A. Streamlined tandem mass tag (SL-TMT) protocol: an efficient strategy for quantitative (phospho)proteome profiling using tandem mass tag-synchronous precursor selection-MS3. J. Proteome Res. 17, 2226–2236 (2018).
McAlister, G. C. et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976–989 (1994).
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Huttlin, E. L. et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010).
Beausoleil, S. A., Villen, J., Gerber, S. A., Rush, J. & Gygi, S. P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 (2006).
Lyons, J. et al. Integrated in vivo multiomics analysis identifies p21-activated kinase signaling as a driver of colitis. Sci. Signal 11, eaan3580 (2018).
Acknowledgements
We thank M. Kostic for critical reading of the manuscript and S. Nabet and members of the Gray laboratory for helpful discussions. We gratefully acknowledge S. Gygi for use of CORE for mass spectrometry data analysis software. This work was supported by an American Cancer Society Postdoctoral Fellowship PF-17-010-01-CDD (B.N.), Claudia Adams Barr Program in Innovative Basic Cancer Research Award (B.N.), Katherine L. and Steven C. Pinard Research Fund (N.S.G. and B.N.), Hope Funds for Cancer Research Postdoctoral Fellowship (S.R.), Harvard Catalyst KL2/CMeRIT Fellowship (S.R.), Perry Levy Fellowship (S.R.), the Lustgarten Foundation (S.R., B.M.W., A.J.A. and W.C.H.), NCI HCMI program (A.J.A. and S.R.), American Cancer Society 129089-PF-16-088-01-TBG (E.J.P.), KU-KIST Graduate School of Converging Science and Technology Program (T.S.), Spanish Ministerio de Economia y Competitividad grant no. SAF2015-60268R, cofunded by Fondo Europeo de Desarrollo Regional funds (J.M.L.), Pancreatic Cancer Action Network Catalyst Award (A.J.A.), Doris Duke Charitable Foundation Clinician Scientist Development Award (A.J.A.), NCI K08 CA218420 (A.J.A.), NCI U01 CA176058 (W.C.H.), NCI U01 CA199253 (W.C.H.) and NCI U01 CA224146 (W.C.H.), American Cancer Society Award 132205-RSG-18-039-01-DMC (K.D.W.), Welch Foundation grant no. I1829 (K.D.W.), 2017 AACR-Bayer Innovation and Discovery grant no. 17-80-44-GRAY (N.S.G.), DF/HCC GI SPORE Developmental Research Project Award P50CA127003 (N.S.G. and K.M.H.) and Hale Center for Pancreatic Research (J.D.M., B.M.W., A.J.A., W.C.H. and N.S.G.).
Author information
Authors and Affiliations
Contributions
F.M.F., B.N. and N.S.G. conceived and led the study. F.M.F. performed medicinal chemistry optimization, with input from J.W. B.N. developed and characterized dTAG model systems, performed cellular PDAC studies and performed DCLK1 nanoBRET assays. B.N., S.R., R.L.K., R.W.S.N. and R.S. performed patient-derived organoid culture assays and prepared organoid samples for RNA-sequencing, proteomics and phosphoproteomics. B.N. and A.L.L. performed pulldown experiments and prepared cellular samples for KiNativ, RNA-sequencing, proteomics and phosphoproteomics. Y.L., W.H. and L.L. performed DCLK1 protein purification, ITC and molecular shift assay. M.K. performed proteomics and phosphoproteomics experiments. A.Y. performed the maximum tolerated dose studies. S.H. performed zebrafish toxicity assays. E.J.P. performed western blot and immunohistochemistry analysis of mouse model tissues. L.H. performed validation of organoid sensitivity. J.K. performed RNA-sequencing data analysis. N.D.-M. and S.E performed ERK5 biochemical and cellular assays. Z.Z., C.R.C., J.D.V. and M.B.R. developed the DCLK1 nanoBRET assay reagents. R.O. performed viability and morphology testing on rat cortical neurons. T.S. and N.D.K. performed molecular modeling. J.M.L., S.M., C.Y.L., A.T.L., K.M.H., J.D.M., B.M.W., A.J.A., W.C.H., K.D.W. and N.S.G. supervised the study. F.M.F., B.N. and S.R. wrote the manuscript, with edits from N.S.G. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
F.M.F. and N.S.G. are inventors on a patent application related to the DCLK1 inhibitors described in this manuscript (WO/2018/075608). B.N. is an inventor on patent applications related to the dTAG system described in this manuscript (WO/2017/024318, WO/2017/024319, WO/2018/148443, WO/2018/148440). Z.Z., C.R.C., J.D.V. and M.B.R. are employees of Promega Corporation. B.M.W. receives research funding from Celgene, Inc, and is a consultant for G1 Therapeutics, BioLineRx and GRAIL. A.J.A. has consulted for Oncorus, Inc. W.C.H. is a consultant for Thermo Fisher, AjuIB, MPM Capital, iTeos and Paraxel and is a Scientific Founder and serves on the Scientific Advisory Board (SAB) for KSQ Therapeutics. K.D.W. is a member of the SAB for Vibliome Therapeutics. N.S.G. is a Scientific Founder, member of the SAB and equity holder in C4 Therapeutics, Syros, Soltego, B2S, Gatekeeper and Petra Pharmaceuticals. The Gray laboratory receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Janssen, Kinogen, Voroni, Her2llc, Deerfield and Sanofi.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Figs. 1–22 and Note.
Supplementary Dataset 1. KinomeSCAN.
Full KINOMEscan dataset.
Supplementary Dataset 2. KiNativ.
Full KiNativ dataset from lysate and live cell format.
Supplementary Dataset 3. RNA-sequencing.
RNA-sequencing dataset for PANFR0172_T2, PANFR0172_T3 and PATU-8988T on comparison of DMSO and 2.5 µM DCLK1-IN-1 treatments for 24 h.
Supplementary Dataset 4. Proteomics.
Mass spectrometry-based proteomics dataset for PANFR0172_T2, PANFR0172_T3 and PATU-8988T on comparison of DMSO and 2.5 µM DCLK1-IN-1 treatments for 24 h.
Supplementary Dataset 5. Phosphoproteomics.
Mass spectrometry-based phosphoproteomics dataset for PANFR0172_T2, PANFR0172_T3 and PATU-8988T on comparison of DMSO and 2.5 µM DCLK1-IN-1 treatments for 24 h.
Rights and permissions
About this article
Cite this article
Ferguson, F.M., Nabet, B., Raghavan, S. et al. Discovery of a selective inhibitor of doublecortin like kinase 1. Nat Chem Biol 16, 635–643 (2020). https://doi.org/10.1038/s41589-020-0506-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0506-0
This article is cited by
-
Crosstalk between colorectal CSCs and immune cells in tumorigenesis, and strategies for targeting colorectal CSCs
Experimental Hematology & Oncology (2024)
-
Applications of lung cancer organoids in precision medicine: from bench to bedside
Cell Communication and Signaling (2023)
-
Macrophage DCLK1 promotes obesity-induced cardiomyopathy via activating RIP2/TAK1 signaling pathway
Cell Death & Disease (2023)
-
Improving the prognosis of pancreatic cancer: insights from epidemiology, genomic alterations, and therapeutic challenges
Frontiers of Medicine (2023)
-
An atlas of substrate specificities for the human serine/threonine kinome
Nature (2023)