Abstract
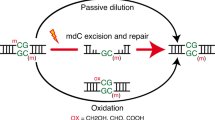
The proposal that N6-methyl-deoxyadenosine (m6dA) acts as an epigenetic mark in mammals remains controversial. Using isotopic labeling coupled to ultrasensitive mass spectrometry, we confirm the presence of low-level m6dA in mammalian DNA. However, the bulk of genomic m6dA originates from ribo-N6-methyladenosine, which is processed via the nucleotide-salvage pathway and misincorporated by DNA polymerases. Our results argue against m6dA acting as a heritable, epigenetic DNA mark in mammalian cells.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information file. Source Data for Figs. 1 and 2 are available online.
References
Wion, D. & Casadesús, J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4, 183–192 (2006).
Koziol, M. J. et al. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 23, 24–30 (2016).
Wu, T. P. et al. DNA methylation on N 6-adenine in mammalian embryonic stem cells. Nature 532, 329–333 (2016).
Xie, Q. et al. N 6-methyladenine DNA modification in glioblastoma. Cell 175, 1228–1243.e20 (2018).
Schiffers, S. et al. Quantitative LC–MS provides no evidence for m6dA or m4dC in the genome of mouse embryonic stem cells and tissues. Angew. Chem. Int. Ed. 6, 11268–11271 (2017).
Ratel, D. et al. Undetectable levels of N6-methyl adenine in mouse DNA: cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett. 580, 3179–3184 (2006).
Liu, B., Liu, X., Lai, W. & Wang, H. Metabolically generated stable isotope-labeled deoxynucleoside code for tracing DNA N6-methyladenine in human cells. Anal. Chem. 89, 6202–6209 (2017).
O’Brown, Z. K. et al. Sources of artifact in measurements of 6mA and 4mC abundance in eukaryotic genomic DNA. BMC Genomics 20, 445 (2019).
Huang, W. et al. Determination of DNA adenine methylation in genomes of mammals and plants by liquid chromatography/mass spectrometry. RSC Adv. 5, 64046–64054 (2015).
Schomacher, L. et al. Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat. Struct. Mol. Biol. 23, 116–124 (2016).
Patil, D. P., Pickering, B. F. & Jaffrey, S. R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28, 113–127 (2018).
Wang, L. Mitochondrial purine and pyrimidine metabolism and beyond. Nucl. Nucl. Nucl. 35, 578–594 (2016).
Löffler, M., Fairbanks, L. D., Zameitat, E., Marinaki, A. M. & Simmonds, H. A. Pyrimidine pathways in health and disease. Trends Mol. Med. 11, 430–437 (2005).
Nyhan, W. L. Nucleotide synthesis via salvage pathway. In Encyclopedia of Life Sciences (John Wiley & Sons, Ltd, 2001).
Charles, M. P. et al. N 6-Methyldeoxyadenosine, a nucleoside commonly found in prokaryotes, induces C2C12 myogenic differentiation. Biochem. Bioph. Res. Co. 314, 476–482 (2004).
Engel, J. D. & Hippel, P. Hvon D.(M6ATP) as a probe of the fidelity of base incorporation into polynucleotides by Escherichia coli DNA polymerase I. J. Biol. Chem. 253, 935–939 (1978).
Chen, M. et al. m6A RNA degradation products are catabolized by an evolutionarily conserved N6-methyl-AMP deaminase in plant and mammalian cells. Plant Cell 30, 1511–1522 (2018).
Xiao, C.-L. et al. N6-methyladenine DNA modification in the human genome. Mol. Cell. 71, 306–318.e7 (2018).
Kweon, S.-M. et al. An adversarial DNA N6-methyladenine-sensor network preserves polycomb silencing. Mol. Cell. 74, 1138–1147.e6 (2019).
Wu, C., Jin, X., Tsueng, G., Afrasiabi, C. & Su, A. I. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 44, D313–D316 (2016).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Yao, B. et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 8, 1122 (2017).
Thomson, J. P. et al. Comparative analysis of affinity-based 5-hydroxymethylation enrichment techniques. Nucleic Acids Res. 41, e206 (2013).
Lentini, A. et al. A reassessment of DNA-immunoprecipitation-based genomic profiling. Nat. Methods 15, 499–504 (2018).
Lane, A. N. & Fan, T. W.-M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 43, 2466–2485 (2015).
Yang, N. C., Ho, W. M., Chen, Y. H. & Hu, M. L. A convenient one-step extraction of cellular ATP using boiling water for the luciferin-luciferase assay of ATP. Anal. Biochem. 306, 323–327 (2002).
Knuckles, P. et al. RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat. Struct. Mol. Biol. 24, 561–569 (2017).
Ruvinsky, A. & Graves, J. A. M. Mammalian Genomics (CABI, 2005).
Acknowledgements
We thank L. Schomacher for discussions, valuable suggestions, including experimental design and feedback during manuscript preparation. We thank M. Pradhan and G. Joshi for discussions, suggestions and critical reading of the manuscript. We are grateful for technical support by S. Ritz from the Institute of Molecular Biology Core Facility Microscopy.
Author information
Authors and Affiliations
Contributions
M.U.M. and C.N. designed the experiments. M.U.M., A.B. and L.K. performed the experiments. C.N. and M.U.M. wrote the manuscript. All authors were involved in discussions, planning the experiments and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Tables 1–7
Source data
Source Data Fig. 1
Source Data Fig. 1c
Source Data Fig. 2
Source Data Fig. 2c–e
Rights and permissions
About this article
Cite this article
Musheev, M.U., Baumgärtner, A., Krebs, L. et al. The origin of genomic N6-methyl-deoxyadenosine in mammalian cells. Nat Chem Biol 16, 630–634 (2020). https://doi.org/10.1038/s41589-020-0504-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0504-2
This article is cited by
-
A novel N6-Deoxyadenine methyltransferase METL-9 modulates C. elegans immunity via dichotomous mechanisms
Cell Research (2023)
-
Navigating the pitfalls of mapping DNA and RNA modifications
Nature Reviews Genetics (2023)
-
Genome-wide deposition of 6-methyladenine in human DNA reduces the viability of HEK293 cells and directly influences gene expression
Communications Biology (2023)
-
Structural insights into DNA N6-adenine methylation by the MTA1 complex
Cell Discovery (2023)
-
Rare and misincorporated DNA N6-methyladenine is a hallmark of cytotoxic stresses for selectively stimulating the stemness and proliferation of glioblastoma cells
Cell Discovery (2022)