Abstract
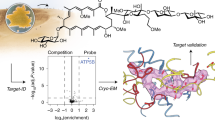
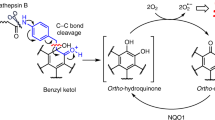
We recently described glutathione peroxidase 4 (GPX4) as a promising target for killing therapy-resistant cancer cells via ferroptosis. The onset of therapy resistance by multiple types of treatment results in a stable cell state marked by high levels of polyunsaturated lipids and an acquired dependency on GPX4. Unfortunately, all existing inhibitors of GPX4 act covalently via a reactive alkyl chloride moiety that confers poor selectivity and pharmacokinetic properties. Here, we report our discovery that masked nitrile-oxide electrophiles, which have not been explored previously as covalent cellular probes, undergo remarkable chemical transformations in cells and provide an effective strategy for selective targeting of GPX4. The new GPX4-inhibiting compounds we describe exhibit unexpected proteome-wide selectivity and, in some instances, vastly improved physiochemical and pharmacokinetic properties compared to existing chloroacetamide-based GPX4 inhibitors. These features make them superior tool compounds for biological interrogation of ferroptosis and constitute starting points for development of improved inhibitors of GPX4.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
References
Viswanathan, V. S. et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017).
Hangauer, M. J. et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 (2017).
Tsoi, J. et al. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell 33, 890–904 (2018).
Yang, W. S. et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014).
Thomas, J. P., Geiger, P. G., Maiorino, M., Ursini, F. & Girotti, A. W. Enzymatic reduction of phospholipid and cholesterol hydroperoxides in artificial bilayers and lipoproteins. Biochim. Biophys. Acta Lipids Lipid Metab. 1045, 252–260 (1990).
Kühn, H. & Borchert, A. Regulation of enzymatic lipid peroxidation: the interplay of peroxidizing and peroxide reducing enzymes. Free Radic. Biol. Med. 33, 154–172 (2002).
Scheerer, P. et al. Structural basis for catalytic activity and enzyme polymerization of phospholipid. Biochemistry 46, 9041–9049 (2007).
Borchert, A. et al. Crystal structure and functional characterization of selenocysteine-containing glutathione peroxidase 4 suggests an alternative mechanism of peroxide reduction. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 1095–1107 (2018).
Yang, W. S. et al. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl Acad. Sci. USA 113, E4966–E4975 (2016).
Shimada, K. et al. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 12, 497–503 (2016).
Yang, W. S. & Stockwell, B. R. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 15, 234–245 (2008).
Sakamoto, K. et al. Discovery of GPX4 inhibitory peptides from random peptide T7 phage display and subsequent structural analysis. Biochem. Biophys. Res. Commun. 482, 195–201 (2017).
Jiang, C., Chen, R., Pandey, A., Kalita, B. & Duraiswamy, A. J. Compounds and method of use. US patent 2019/0263802 A1. 1–292 (2019).
Weïwer, M. et al. Development of small-molecule probes that selectively kill cells induced to express mutant RAS. Bioorg. Med. Chem. Lett. 22, 1822–1826 (2012).
Seashore-Ludlow, B. et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Disco. 5, 1210–1223 (2015).
Rees, M. G. et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 12, 109–116 (2015).
Basu, A. et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–1161 (2013).
Gaschler, M. M. et al. FINO2 initiates ferroptosis through GPX4 inactivation and iron oxidation. Nat. Chem. Biol. 14, 507–515 (2018).
Dixon, S. J. et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Skouta, R. et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J. Am. Chem. Soc. 136, 4551–4556 (2014).
Zilka, O. et al. On the mechanism of cytoprotection by ferrostatin-1 and liproxstatin-1 and the role of lipid peroxidation in ferroptotic cell death. ACS Cent. Sci. 3, 232–243 (2017).
Molina, D. M. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Gao, J. et al. Selenium-encoded isotopic signature targeted profiling. ACS Cent. Sci. 4, 960–970 (2018).
Friedmann Angeli, J. P. et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191 (2014).
Trefzer, C. et al. Benzothiazinones: prodrugs that covalently modify the decaprenylphosphoryl-beta-d-ribose 2’-epimerase DprE1 of Mycobacterium tuberculosis. J. Am. Chem. Soc. 132, 13663–13665 (2010).
Patterson, S. & Wyllie, S. Nitro drugs for the treatment of trypanosomatid diseases: past, present, and future prospects. Trends Parasitol. 30, 289–298 (2014).
Yu, J. et al. Elucidation of a novel bioactivation pathway of a 3,4-unsubstituted isoxazole in human liver microsomes: formation of a glutathione adduct of a cyanoacrolein derivative after isoxazole ring opening. Drug Metab. Dispos. 39, 302–311 (2011).
Duranti, E., Balsamini, C., Spadoni, G. & Staccioli, L. Reaction of secondary acetylenic bromides with sodium nitrite: synthesis of 3,5-alkyl(aryl)-4-nitroisoxazoles. J. Org. Chem. 53, 2870–2872 (1988).
Ray, S., Kreitler, D. F., Gulick, A. M. & Murkin, A. S. The nitro group as a masked electrophile in covalent enzyme inhibition. ACS Chem. Biol. 13, 1470–1473 (2018).
Curini, M. et al. Alumina promoted cyclization of α-nitro-oximes: a new entry to the synthesis of 1,2,5-oxadiazoles N-oxides (furoxans). Tetrahedron Lett. 41, 8817–8820 (2000).
Zhao, J. Q. et al. Synthesis of furoxan derivatives: DABCO-mediated cascade sulfonylation/cyclization reaction of α-nitro-ketoximes. Tetrahedron 71, 1560–1565 (2015).
Burakevich, J. V., Butler, R. S. & Volpp, G. P. Phenylfurazan oxide. Chemistry. J. Org. Chem. 37, 593–596 (1972).
Kalinina, M. I. & Mosiev, I. K. Properties of furoxans monosubstituted with adamantanes. Chem. Heterocycl. Compd. 24, 217–220 (1988).
Eaton, J. K., Ruberto, R. A., Kramm, A., Viswanathan, V. S. & Schreiber, S. L. Diacylfuroxans are masked nitrile oxides that inhibit GPX4 covalently. J. Am. Chem. Soc. 141, 20407–20415 (2019).
Shelton, B. R., Howe, R. & Liu, K. C. A particularly convenient preparation of benzohydroximinoyl chlorides (nitrile oxide precursors). J. Org. Chem. 45, 3916–3918 (1980).
Egan, C., Clery, M., Hegarty, A. F. & Welch, A. J. Mechanism of reaction of isomeric nitrolic acids to nitrile oxides in aqueous solution. J. Chem. Soc. Perkin Trans. 2, 249–256 (1991).
Matt, C., Gissot, A., Wagner, A. & Mioskowski, C. Nitrolic acids: efficient precursors of nitrile oxides under neutral conditions. Tetrahedron Lett. 41, 1191–1194 (2000).
Matt, C., Wagner, A. & Mioskowski, C. Novel transformation of primary nitroalkanes and primary alkyl bromides to the corresponding carboxylic acids. J. Org. Chem. 62, 234–235 (1997).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
Shah, R., Shchepinov, M. S. & Pratt, D. A. Resolving the role of lipoxygenases in the initiation and execution of ferroptosis. ACS Cent. Sci. 4, 387–396 (2018).
Weerapana, E. et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–797 (2010).
Backus, K. M. et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016).
Bar-Peled, L. et al. Chemical proteomics identifies druggable vulnerabilities in a genetically defined cancer. Cell 171, 696–709 (2017).
Allimuthu, D. & Adams, D. J. 2-Chloropropionamide as a low-reactivity electrophile for irreversible small-molecule probe identification. ACS Chem. Biol. 12, 2124–2131 (2017).
Shindo, N. et al. Selective and reversible modification of kinase cysteines with chlorofluoroacetamides. Nat. Chem. Biol. 15, 250–258 (2019).
Grundmann, C. & Dean, J. M. Nitrile oxides. V. Stable aromatic nitrile oxides. J. Org. Chem. 30, 2809–2812 (1965).
Zaro, B. W., Whitby, L. R., Lum, K. M. & Cravatt, B. F. Metabolically labile fumarate esters impart kinetic selectivity to irreversible inhibitors. J. Am. Chem. Soc. 138, 15841–15844 (2016).
Martín-Gago, P. et al. Covalent protein labeling at glutamic acids. Cell Chem. Biol. 24, 589–597.e5 (2017).
Geu-Flores, F. et al. Glucosinolate engineering identifies a γ-glutamyl peptidase. Nat. Chem. Biol. 5, 575–577 (2009).
Mutlib, A.E. et al. P450-mediated metabolism of 1-[3-(aminomethyl)phenyl]-N-[3-fluoro-2’-(methylsulfonyl)-[1,1’-biphenyl]-4-yl]-3-(trifluoromethyl)-1H-pyrazole-5-carboxamide (DPC 423) and its analogues to aldoximes. Characterization of glutathione conjugates of postulated intermediates derived from aldoximes. Chem. Res. Toxicol. 15, 63–75 (2002).
Roveri, A., Maiorino, M. & Ursini, F. Enzymatic and immunological measurements of soluble and membrane-bound phospholipid-hydroperoxide glutathione peroxidase. Methods Enzymol. 233, 202–212 (1994).
Kato, S. et al. Preparation of 13 or 9-hydroperoxy-9Z,11E (9E,11E) or 10E,12Z (10E,12E)-octadecadienoic phosphatidylcholine hydroperoxide. J. Oleo Sci. 63, 431–437 (2014).
Kriska, T. & Girotti, A. W. A thin layer chromatographic method for determining the enzymatic activity of peroxidases catalyzing the two-electron reduction of lipid hydroperoxides. J. Chromatogr. B. 827, 58–64 (2005).
Novoselov, S. V. et al. A highly efficient form of the selenocysteine insertion sequence element in protozoan parasites and its use in mammalian cells. Proc. Natl Acad. Sci. USA 104, 7857–7862 (2007).
Durocher, Y., Perret, S. & Kamen, A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30, e9 (2002).
Nguyen, D. et al. Discovery and characterization of the potent and highly selective(piperidin-4-yl)pyrido[3,2-d]pyrimidine based in vitro probe BAY-885 for the kinase ERK5. J. Med. Chem. 62, 928–940 (2019).
Werner, S. et al. Discovery and characterization of the potent and selective P2X4 inhibitor N-[4-(3-chlorophenoxy)-3-sulfamoylphenyl]-2-phenylacetamide (BAY-1797) and structure-guided amelioration of its CYP3A4 induction profile. J. Med. Chem. 62, 11194–11217 (2019).
Cee, V. J. et al. Systematic study of the glutathione (GSH) reactivity of N-arylacrylamides: 1. effects of aryl substitution. J. Med. Chem. 58, 9171–9178 (2015).
Acknowledgements
We thank V. Kaushik for assistance with intact protein mass spectrometry experiments; M. Palte for assistance with preparing phosphatidylcholine hydroperoxide; B. Budnik and R. Robinson for assistance with proteomics. This work was supported in part by the National Institute of General Medical Sciences (R01GM038627 and R35GM127045 awarded to S.L.S.) and through a collaboration between the Broad Institute and Bayer AG.
Author information
Authors and Affiliations
Contributions
J.K.E. conceived and designed experiments. J.K.E., L.F., K.E.L., S.G. and C.M. performed chemical synthesis and compound characterization. J.K.E., R.A.R., M.J.R., L.L.C. and V.S.V. maintained cell cultures and performed viability, cellular thermal shift and western blotting experiments. D.M. and A.H. designed the cloning approach and expressed, purified and characterized recombinant wild-type GPX4 protein. V.B., A.H., D.M. and J.K.E. performed cellular and biochemical mass spectrometry binding assays. M.N. performed metabolite-ID studies. R.C.H., K.Z., A.K., S. Chen and B.B. contributed tools and reagents for protein characterization experiments. P.A.C. contributed to analysis of cell viability data. R.A.R. and S. Christian performed cellular lipid peroxidation assays. R.N. performed formulation work. A.L.E. performed in vivo experiments. J.K.E., V.S.V. and S.L.S. initiated the project and wrote the manuscript. V.S.V. and S.L.S. directed the project.
Corresponding authors
Ethics declarations
Competing interests
S.L.S. serves on the Board of Directors of the Genomics Institute of the Novartis Research Foundation (“GNF”); is a shareholder and serves on the Board of Directors of Jnana Therapeutics; is a shareholder of Forma Therapeutics; is a shareholder and advises Kojin Therapeutics, Kisbee Therapeutics, Decibel Therapeutics and Eikonizo Therapeutics; serves on the Scientific Advisory Boards of Eisai Co., Ltd., Ono Pharma Foundation, Exo Therapeutics, and F-Prime Capital Partners; and is a Novartis Faculty Scholar. P.A.C. is an advisor to Pfizer, Inc. D.M., A.H., K.Z., M.N., V.B., R.C.H., S.G., S. Christian, R.N. and A.L.E. are employed by Bayer AG.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–3, Figs. 1–19 and synthetic methods.
Rights and permissions
About this article
Cite this article
Eaton, J.K., Furst, L., Ruberto, R.A. et al. Selective covalent targeting of GPX4 using masked nitrile-oxide electrophiles. Nat Chem Biol 16, 497–506 (2020). https://doi.org/10.1038/s41589-020-0501-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0501-5
This article is cited by
-
TrkA promotes MDM2-mediated AGPS ubiquitination and degradation to trigger prostate cancer progression
Journal of Experimental & Clinical Cancer Research (2024)
-
Synergism of non-thermal plasma and low concentration RSL3 triggers ferroptosis via promoting xCT lysosomal degradation through ROS/AMPK/mTOR axis in lung cancer cells
Cell Communication and Signaling (2024)
-
Low-dose hypomethylating agents cooperate with ferroptosis inducers to enhance ferroptosis by regulating the DNA methylation-mediated MAGEA6-AMPK-SLC7A11-GPX4 signaling pathway in acute myeloid leukemia
Experimental Hematology & Oncology (2024)
-
CGI1746 targets σ1R to modulate ferroptosis through mitochondria-associated membranes
Nature Chemical Biology (2024)
-
Mitochondrial regulation of GPX4 inhibition–mediated ferroptosis in acute myeloid leukemia
Leukemia (2024)