Abstract
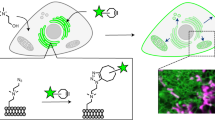
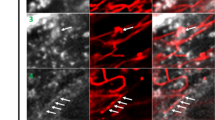
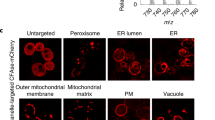
We report new lipid-based, high-density, environmentally sensitive (HIDE) probes that accurately and selectively image endo-lysosomes and their dynamics at super-resolution for extended times. Treatment of live cells with the small molecules DiIC16TCO or DiIC16’TCO followed by in situ tetrazine ligation reaction with the silicon-rhodamine dye SiR-Tz generates the HIDE probes DiIC16-SiR and DiIC16’-SiR in the endo-lysosomal membrane. These new probes support the acquisition of super-resolution videos of organelle dynamics in primary cells for more than 7 min with no detectable change in endosome structure or function. Using DiIC16-SiR and DiIC16’-SiR, we describe direct evidence of endosome motility defects in cells from patients with Niemann–Pick Type-C disease. In wild-type fibroblasts, the probes reveal distinct but rare inter-endosome kiss-and-run events that cannot be observed using confocal methods. Our results shed new light on the role of NPC1 in organelle motility and cholesterol trafficking.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The materials and data reported in this study are available upon reasonable request from the corresponding author.
References
Maxfield, F. R. & McGraw, T. E. Endocytic recycling. Nat. Rev. Mol. Cell Bio. 5, 121–132 (2004).
Bonifacino, J. S. & Neefjes, J. Moving and positioning the endolysosomal system. Curr. Opin. Cell Biol. 47, 1–8 (2017).
Sadowski, L., Pilecka, I. & Miaczynska, M. Signaling from endosomes: location makes a difference. Exp. Cell. Res. 315, 1601–1609 (2009).
Alabi, A. A. & Tsien, R. W. Perspectives on kiss-and-run: role in exocytosis, endocytosis, and neurotransmission. Annu Rev. Physiol. 75, 393–422 (2013).
Gruenberg, J. The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell Bio. 2, 721–730 (2001).
Li, D. et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349, 6251 (2015).
Shim, S. H. et al. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl Acad. Sci. USA 109, 13978–13983 (2012).
Richardson, D. S. et al. SRpHi ratiometric pH biosensors for superresolution microscopy. Nat. Commun. 8, 577 (2017).
Wegel, E. et al. Imaging cellular structures in super-resolution with SIM, STED and localisation microscopy: a practical comparison. Sci. Rep. 6, 27290 (2016).
Das, A. & Barroso, M. M. Endosome-mitochondria interactions are modulated by iron release from transferrin. Mol. Biol. Cell 27, 831–845 (2016).
Ba, Q., Raghavan, G., Kiselyov, K. & Yang, G. Whole-cell scale dynamic organization of lysosomes revealed by spatial statistical analysis. Cell Rep. 23, 3591–3606 (2018).
Platt, F. M., Boland, B. & van der Spoel, A. C. Lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J. Cell Biol. 199, 723–734 (2012).
Futerman, A. H. & van Meer, G. The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Bio. 5, 554–565 (2004).
Pfeffer, S. R. NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J. Biol. Chem. 294, 1706–1709 (2019).
Lim, C. Y. et al. ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann–Pick type C. Nat. Cell Biol. 21, 1206–1218 (2019).
Leung, K., Chakraborty, K., Saminathan, A., Krishnan, Y. & DNA, A. nanomachine chemically resolves lysosomes in live cells. Nat. Nanotechnol. 14, 176–183 (2019).
Vacca, F. et al. Cyclodextrin triggers MCOLN1-dependent endo-lysosome secretion in Niemann–Pick type C cells. J. Lipid Res. 60, 832–843 (2019).
Vivas, O. et al. Disease reveals a link between lysosomal cholesterol and PtdIns(4,5)P-2 that regulates neuronal excitability. Cell Rep. 27, 2636–2648 (2019).
Vanier, M. T. & Latour, P. Laboratory diagnosis of Niemann–Pick disease type C: the filipin staining test. Method Cell Biol. 126, 357–375 (2015).
Gelsthorpe, M. E. et al. Niemann–Pick type C1I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J. Biol. Chem. 283, 8229–8236 (2008).
Deffieu, M. S. & Pfeffer, S. R. Niemann–Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc. Natl Acad. Sci. USA 108, 18932–18936 (2011).
Park, W. D. et al. Identification of 58 novel mutations in Niemann–Pick disease Type C: correlation with biochemical phenotype and importance of PTC1-like domains in NPC1. Hum. Mutat. 22, 313–325 (2003).
Lu, F. et al. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife 4, e12177 (2015).
Ko, D. C., Gordon, M. D., Jin, J. Y. & Scott, M. P. Dynamic movements of organelles containing Niemann–Pick C1 protein: NPC1 involvement in late endocytic events. Mol. Biol. Cell 12, 601–614 (2001).
Millat, G. et al. Niemann–Pick C1 disease: correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol-sensing domain and of the cysteine-rich luminal loop. Am. J. Hum. Genet. 68, 1373–1385 (2001).
Lebrand, C. et al. Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J. 21, 1289–1300 (2002).
Sobo, K. et al. Late endosomal cholesterol accumulation leads to impaired intra-endosomal trafficking. PLoS ONE 2, e851 (2007).
Thompson, A. D., Bewersdorf, J., Toomre, D. & Schepartz, A. HIDE Probes: a new toolkit for visualizing organelle dynamics, longer and at super-resolution. Biochem. 56, 5194–5201 (2017).
Erdmann, R. S. et al. Super-resolution imaging of the Golgi in live cells with a bioorthogonal ceramide probe. Angew. Chem. Int. Ed. 53, 10242–10246 (2014).
Takakura, H. et al. Long time-lapse nanoscopy with spontaneously blinking membrane probes. Nat. Biotechnol. 35, 773–780 (2017).
Bottanelli, F. et al. A novel physiological role for ARF1 in the formation of bidirectional tubules from the Golgi. Mol. Biol. Cell 28, 1676–1687 (2017).
Bottanelli, F. et al. Two-colour live-cell nanoscale imaging of intracellular targets. Nat. Commun. 7, 10778 (2016).
Revelo, N. H. et al. A new probe for super-resolution imaging of membranes elucidates trafficking pathways. J. Cell Biol. 205, 591–606 (2014).
Blackman, M. L., Royzen, M. & Fox, J. M. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels–Alder reactivity. J. Am. Chem. Soc. 130, 13518–13519 (2008).
Paz, I. et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 12, 530–544 (2010).
Thurston, T. L. M., Wandel, M. P., von Muhlinen, N., Foeglein, A. & Randow, F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–U1515 (2012).
Thompson, A. D. et al. Long-term live-cell STED nanoscopy of primary and cultured cells with the plasma membrane HIDE Probe DiI-SiR. Angew. Chem., Int. Ed. Engl. 56, 10408–10412 (2017).
Sun, X. F. et al. Niemann–Pick C variant detection by altered sphingolipid trafficking and correlation with mutations within a specific domain of NPC1. Am. J. Hum. Genet. 68, 1361–1372 (2001).
Salman, A., Cougnoux, A., Farhat, N., Wassif, C. A. & Porter, F. D. Association of NPC1 Variant p. P237S with a pathogenic splice variant in two Niemann–Pick disease type C1 patients. Am. J. Med Genet. A. 173, 1038–1040 (2017).
Pipalia, N. H. et al. Histone deacetylase inhibitors correct the cholesterol storage defect in most Niemann–Pick C1 mutant cells. J. Lipid Res. 58, 695–708 (2017).
Li, X. C., Saha, P., Li, J., Blobel, G. & Pfeffer, S. R. Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc. Natl Acad. Sci. USA 113, 10079–10084 (2016).
Eden, E. R. The formation and function of ER-endosome membrane contact sites. BBA-Mol. Cell Biol. L. 1861, 874–879 (2016).
Raiborg, C., Wenzel, E. M. & Stenmark, H. ER-endosome contact sites: molecular compositions and functions. EMBO J. 34, 1848–1858 (2015).
Watanabe, S. et al. Ultrafast endocytosis at mouse hippocampal synapses. Nature 504, 242–247 (2013).
Staal, R. G. W., Mosharov, E. V. & Sulzer, D. Dopamine neurons release transmitter via a flickering fusion pore. Nat. Neurosci. 7, 341–346 (2004).
Saffi, G. T. & Botelho, R. J. Lysosome fission: planning for an exit. Trends Cell Biol. 29, 635–646 (2019).
Vihervaara, T. et al. Sterol binding by OSBP-related protein 1L (ORP1 L) regulates late endosome motility and function. Cell. Mol. Life Sci. 68, 537–551 (2011).
Chen, H., Yang, J., Low, P. S. & Cheng, J. X. Cholesterol level regulates endosome motility via Rab proteins. Biophys. J. 94, 1508–1520 (2008).
Millard, E. E. et al. The sterol-sensing domain of the Niemann–Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J. Biol. Chem. 280, 28581–28590 (2005).
Li, X. et al. 3.3 A structure of Niemann–Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport. Proc. Natl Acad. Sci. USA 114, 9116–9121 (2017).
Mukherjee, S. & Maxfield, F. R. Trafficking of lipid analogs with varying tail lengths and unsaturations in Niemann–Pick C fibroblasts. Mol. Biol. Cell 11, 325a–325a (2000).
Devaraj, N. K., Upadhyay, R., Hatin, J. B., Hilderbrand, S. A. & Weissleder, R. Fast and sensitive pretargeted labeling of cancer cells through a tetrazine/trans-cyclooctene cycloaddition. Angew. Chem. Int. Ed. 48, 7013–7016 (2009).
Roberts, R. L., Barbieri, M. A., Pryse, K. M., Chua, M. & Stahl, P. D. Endosome fusion in living cells overexpressing GFP-rab5. J. Cell Sci. 112, 3667–3675 (1999).
Humphries, W. H., Szymanski, C. J. & Payne, C. K. Endo-lysosomal vesicles positive for Rab7 and LAMP1 are terminal vesicles for the transport of dextran. PLoS ONE 6, e26626 (2011).
Steinauer, A. et al. HOPS-dependent endosomal fusion required for efficient cytosolic delivery of therapeutic peptides and small proteins. Proc. Natl Acad. Sci. USA 116, 512–521 (2019).
Aits, S. et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11, 1408–1424 (2015).
Ibach, J. et al. Single particle tracking reveals that egfr signaling activity is amplified in clathrin-coated pits. PLoS ONE 10, e0143162 (2015).
Roepstorff, K. et al. Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic 10, 1115–1127 (2009).
Tinevez, J. Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Acknowledgements
This work was supported by the NIH (grant nos. R01GM131372-01, A.S. and R01GM118486, D.T.) and the Wellcome Trust (grant no. 095927/A/11/Z), and in part by the NIH (grant no. S10 OD020142 (Leica SP8)). A.G. was in part supported by the NIH (5T32GM06754 3-12). A.G. thanks J. Wolenksi and A. Mennone for assistance with confocal and STED microscopy.
Author information
Authors and Affiliations
Contributions
A.G., D.T., and A.S. conceived the project. A.G., F.R.-M, Z.X., D.T., and A.S. designed experiments. A.G. and F.R.-M. performed imaging experiments. A.G. designed and synthesized DiIC16TCO and DiIC16’TCO. A.G., F.R.-M., and Z.X. prepared HeLa and fibroblast samples for microscopy. A.G. and A.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Note.
Supplementary Video 1
HeLa cells labeled with DiIC16-SiR.
Supplementary Video 2
HeLa cells labeled with DiIC16’-SiR.
Supplementary Video 3
Wild-type (GM05399) fibroblasts labeled with DiIC16-SiR.
Supplementary Video 4
I1061T (GM18453) Fibroblasts labeled with DiIC16-SiR.
Supplementary Video 5
P237S/I1061T (GM03123) Fibroblasts labeled with DiIC16-SiR.
Supplementary Video 6
R404Q (GM18388) Fibroblasts labeled with DiIC16-SiR.
Supplementary Video 7
1920delG (GM23945) Fibroblasts labeled with DiIC16-SiR.
Rights and permissions
About this article
Cite this article
Gupta, A., Rivera-Molina, F., Xi, Z. et al. Endosome motility defects revealed at super-resolution in live cells using HIDE probes. Nat Chem Biol 16, 408–414 (2020). https://doi.org/10.1038/s41589-020-0479-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0479-z
This article is cited by
-
Long-term super-resolution inner mitochondrial membrane imaging with a lipid probe
Nature Chemical Biology (2024)
-
Bioorthogonal Chemistry in Cellular Organelles
Topics in Current Chemistry (2024)
-
Organelle membrane-specific chemical labeling and dynamic imaging in living cells
Nature Chemical Biology (2020)