Abstract
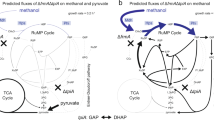
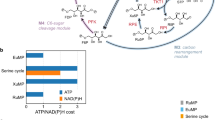
Engineering a biotechnological microorganism for growth on one-carbon intermediates, produced from the abiotic activation of CO2, is a key synthetic biology step towards the valorization of this greenhouse gas to commodity chemicals. Here we redesign the central carbon metabolism of the model bacterium Escherichia coli for growth on one-carbon compounds using the reductive glycine pathway. Sequential genomic introduction of the four metabolic modules of the synthetic pathway resulted in a strain capable of growth on formate and CO2 with a doubling time of ~70 h and growth yield of ~1.5 g cell dry weight (gCDW) per mol-formate. Short-term evolution decreased doubling time to less than 8 h and improved biomass yield to 2.3 gCDW per mol-formate. Growth on methanol and CO2 was achieved by further expression of a methanol dehydrogenase. Establishing synthetic formatotrophy and methylotrophy, as demonstrated here, paves the way for sustainable bioproduction rooted in CO2 and renewable energy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Complete information on the experimental setup as well as detailed results are available from the corresponding author upon reasonable request.
Code availability
MATLAB code used for the analysis of the experiments is available from the corresponding author upon request.
References
Blankenship, R. E. et al. Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science 332, 805–809 (2011).
Scheffe, J. R. & Steinfeld, A. Oxygen exchange materials for solar thermochemical splitting of H2O and CO2: a review. Mater. Today 17, 341–348 (2014).
Snoeckx, R. & Bogaerts, A. Plasma technology—a novel solution for CO2 conversion? Chem. Soc. Rev. 46, 5805–5863 (2017).
Zhang, Q., Kang, J. & Wang, Y. Development of novel catalysts for Fischer–Tropsch synthesis: tuning the product selectivity. ChemCatChem 2, 1030–1058 (2010).
Jouny, M., Luc, W. & Jiao, F. General techno-economic analysis of CO2 electrolysis systems. Ind. Eng. Chem. Res. 57, 2165–2177 (2018).
Yishai, O., Lindner, S. N., Gonzalez de la Cruz, J., Tenenboim, H. & Bar-Even, A. The formate bio-economy. Curr. Opin. Chem. Biol. 35, 1–9 (2016).
Szima, S. & Cormos, C. C. Improving methanol synthesis from carbon-free H2 and captured CO2: a techno-economic and environmental evaluation. J. CO2 Util. 24, 555–563 (2018).
Bertsch, J. & Muller, V. Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria. Biotechnol. Biofuels 8, 210 (2015).
Bennett, R. K., Steinberg, L. M., Chen, W. & Papoutsakis, E. T. Engineering the bioconversion of methane and methanol to fuels and chemicals in native and synthetic methylotrophs. Curr. Opin. Biotechnol. 50, 81–93 (2017).
Muller, J. E. et al. Engineering Escherichia coli for methanol conversion. Metab. Eng. 28, 190–201 (2015).
Dai, Z. et al. Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae. Bioresour. Technol. 245, 1407–1412 (2017).
Yu, H. & Liao, J. C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds. Nat. Commun. 9, 3992 (2018).
Meyer, F. et al. Methanol-essential growth of Escherichia coli. Nat. Commun. 9, 1508 (2018).
Woolston, B. M., King, J. R., Reiter, M., Van Hove, B. & Stephanopoulos, G. Improving formaldehyde consumption drives methanol assimilation in engineered E. coli. Nat. Commun. 9, 2387 (2018).
Bennett, R. K., Gonzalez, J. E., Whitaker, W. B., Antoniewicz, M. R. & Papoutsakis, E. T. Expression of heterologous non-oxidative pentose phosphate pathway from Bacillus methanolicus and phosphoglucose isomerase deletion improves methanol assimilation and metabolite production by a synthetic Escherichia coli methylotroph. Metab. Eng. 45, 75–85 (2017).
Gonzalez, J., Bennett, R. K., Papoutsakis, E. T. & Antoniewicz, M. R. Methanol assimilation in Escherichia coli is improved by co-utilization of threonine and deletion of leucine-responsive regulatory protein. Metab. Eng. 45, 67–74 (2017).
Rohlhill, J., Sandoval, N. R. & Papoutsakis, E. T. Sort-Seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol. ACS Synth. Biol. 6, 1584–1595 (2017).
Woolston, B. M., Roth, T., Kohale, I., Liu, D. R. & Stephanopoulos, G. Development of a formaldehyde biosensor with application to synthetic methylotrophy. Biotechnol. Bioeng. 115, 206–215 (2018).
Whitaker, W. B. et al. Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli. Metab. Eng. 39, 49–59 (2017).
Lu, X. et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design. Nat. Commun. 10, 1378 (2019).
Wang, X. et al. Biological conversion of methanol by evolved Escherichia coli carrying a linear methanol assimilation pathway. Bioresour. Bioprocess. 4, 41–46 (2017).
Anthony, C. The Biochemistry of Methylotrophs (Academic Press, 1982).
Drake, H. L., Kirsten, K. & Matthies, C. Acetogenic Prokaryotes. in The Prokaryotes (eds., Stanley Falkow, Eugene Rosenberg, Karl-Heinz Schleifer, Erko Stackebrandt) 354–420 (Springer, 2006).
Bar-Even, A., Noor, E., Flamholz, A. & Milo, R. Design and analysis of metabolic pathways supporting formatotrophic growth for electricity-dependent cultivation of microbes. Biochim. Biophys. Acta 1827, 1039–1047 (2013).
Bar-Even, A. Does acetogenesis really require especially low reduction potential? Biochim. Biophys. Acta 1827, 395–400 (2013).
Noor, E. et al. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput. Biol. 10, e1003483 (2014).
Figueroa, I. A. et al. Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc. Natl Acad. Sci. USA 115, E92–E101 (2018).
Kawasaki, H., Sato, T. & Kikuchi, G. A new reaction for glycine biosynthesis. Biochem. Biophys. Res. Commun. 23, 227–233 (1966).
Motokawa, Y. & Kikuchi, G. Glycine metabolism by rat liver mitochondria. Reconstruction of the reversible glycine cleavage system with partially purified protein components. Arch. Biochem. Biophys. 164, 624–633 (1974).
Pasternack, L. B., Laude, D. A. Jr. & Appling, D. R. 13C NMR detection of folate-mediated serine and glycine synthesis in vivo in Saccharomyces cerevisiae. Biochemistry 31, 8713–8719 (1992).
Tashiro, Y., Hirano, S., Matson, M. M., Atsumi, S. & Kondo, A. Electrical-biological hybrid system for CO2 reduction. Metab. Eng. 47, 211–218 (2018).
Yishai, O., Bouzon, M., Doring, V. & Bar-Even, A. In vivo assimilation of one-carbon via a synthetic reductive glycine pathway in Escherichia coli. ACS Synth. Biol. 7, 2023–2028 (2018).
Crowther, G. J., Kosaly, G. & Lidstrom, M. E. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J. Bacteriol. 190, 5057–5062 (2008).
Tishkov, V. I. & Popov, V. O. Catalytic mechanism and application of formate dehydrogenase. Biochem. (Mosc.) 69, 1252–1267 (2004).
Wenk, S., Yishai, O., Lindner, S. N. & Bar-Even, A. An engineering approach for rewiring microbial metabolism. Methods Enzymol. 608, 329–367 (2018).
Bassalo, M. C. et al. Rapid and efficient one-step metabolic pathway integration in E. coli. ACS Synth. Biol. 5, 561–568 (2016).
Gleizer, S. et al. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell 179, 1255–1263.e12 (2019).
Claassens, N. J., Cotton, C. A., Kopljar, D. & Bar-Even, A. Making quantitative sense of electromicrobial production. Nat. Catal. 2, 437 (2019).
Nicholls, P. Formate as an inhibitor of cytochrome c oxidase. Biochem. Biophys. Res. Commun. 67, 610–616 (1975).
Warnecke, T. & Gill, R. T. Organic acid toxicity, tolerance, and production in Escherichia coli biorefining applications. Micro. Cell Fact. 4, 25 (2005).
Dragosits, M. & Mattanovich, D. Adaptive laboratory evolution—principles and applications for biotechnology. Micro. Cell Fact. 12, 64 (2013).
Wytock, T. P. et al. Experimental evolution of diverse Escherichia coli metabolic mutants identifies genetic loci for convergent adaptation of growth rate. PLoS Genet. 14, e1007284 (2018).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
Gutheil, W. G., Kasimoglu, E. & Nicholson, P. C. Induction of glutathione-dependent formaldehyde dehydrogenase activity in Escherichia coli and Hemophilus influenza. Biochem. Biophys. Res. Commun. 238, 693–696 (1997).
Kotrbova-Kozak, A., Kotrba, P., Inui, M., Sajdok, J. & Yukawa, H. Transcriptionally regulated adhA gene encodes alcohol dehydrogenase required for ethanol and n-propanol utilization in Corynebacterium glutamicum R. Appl. Microbiol. Biotechnol. 76, 1347–1356 (2007).
Wu, T. Y. et al. Characterization and evolution of an activator-independent methanol dehydrogenase from Cupriavidus necator N-1. Appl. Microbiol. Biotechnol. 100, 4969–4983 (2016).
Roth, T. B., Woolston, B. M., Stephanopoulos, G. & Liu, D. R. Phage-assisted evolution of Bacillus methanolicus methanol dehydrogenase 2. ACS Synth. Biol. 8, 796–806 (2019).
Zhang, W. et al. Expression, purification, and characterization of formaldehyde dehydrogenase from Pseudomonas aeruginosa. Protein Expr. Purif. 92, 208–213 (2013).
Cotton, C. A., Claassens, N. J., Benito-Vaquerizo, S. & Bar-Even, A. Renewable methanol and formate as microbial feedstocks. Curr. Opin. Biotechnol. 62, 168–180 (2020).
Thoma, S. & Schobert, M. An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol. Lett. 294, 127–132 (2009).
Thomason, L. C., Costantino, N. & Court, D. L. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. https://doi.org/10.1002/0471142727.mb0117s79 (2007).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006–2008 (2006).
Nyerges, A. et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc. Natl Acad. Sci. USA 113, 2502–2507 (2016).
Zelcbuch, L. et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 41, e98 (2013).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual 3rd edn. (Cold Spring Harbor Laboratory Press, 2001).
Braatsch, S., Helmark, S., Kranz, H., Koebmann, B. & Jensen, P. R. Escherichia coli strains with promoter libraries constructed by Red/ET recombination pave the way for transcriptional fine-tuning. Biotechniques 45, 335–337 (2008).
Giavalisco, P. et al. Elemental formula annotation of polar and lipophilic metabolites using 13C, 15N and 34S isotope labelling, in combination with high‐resolution mass spectrometry. Plant J. 68, 364–376 (2011).
Liu, A., Feng, R. & Liang, B. Microbial surface displaying formate dehydrogenase and its application in optical detection of formate. Enzym. Microb. Technol. 91, 59–65 (2016).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408 (2001).
Zhou, K. et al. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 12, 18 (2011).
Acknowledgements
We thank C. Cotton, N. Claassens, H. He, R. Milo, E. Noor, N. Antonovsky, A. Flamholz, Y. Bar-On, W. Newell, D. Bonder, A. Satanowski, T. Erb and M. Bouzon for helpful discussions and suggestions. This work was funded by the Max Planck Society, by the German Ministry of Education and Research grant FormatPlant (part of BioEconomy 2030, Plant Breeding Research for the Bioeconomy) and by the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 763911 (Project eForFuel).
Author information
Authors and Affiliations
Contributions
A.B.-E. designed and supervised the research and wrote the paper. S.K., S.N.L., S.A. and O.Y. genetically engineered E. coli for growth on formate and methanol, and performed the growth experiments. S.K. and S.N.L. measured biomass yield on formate and methanol. S.A. performed the qPCR experiments. S.W. and K.S. cloned the methanol dehydrogenase and formaldehyde dehydrogenase genes, and assisted in the growth experiments on methanol. S.K., S.N.L., S.A., O.Y., S.W., K.S. and A.B.-E. analyzed the data.
Corresponding author
Ethics declarations
Competing interests
A.B.-E. is cofounder of b.fab, exploring the commercialization of microbial bioproduction using formate as feedstock. The company was not involved in any way in performing or funding this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1, Figs. 1–12 and Note.
Rights and permissions
About this article
Cite this article
Kim, S., Lindner, S.N., Aslan, S. et al. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat Chem Biol 16, 538–545 (2020). https://doi.org/10.1038/s41589-020-0473-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0473-5
This article is cited by
-
Physiological versatility of ANME-1 and Bathyarchaeotoa-8 archaea evidenced by inverse stable isotope labeling
Microbiome (2024)
-
Synthetic microbiology in sustainability applications
Nature Reviews Microbiology (2024)
-
Prospects of formamide as nitrogen source in biotechnological production processes
Applied Microbiology and Biotechnology (2024)
-
Engineering a synthetic energy-efficient formaldehyde assimilation cycle in Escherichia coli
Nature Communications (2023)
-
Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo
Nature Communications (2023)