Abstract
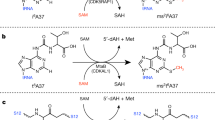
Tryptophan 2C methyltransferase (TsrM) methylates C2 of the indole ring of l-tryptophan during biosynthesis of the quinaldic acid moiety of thiostrepton. TsrM is annotated as a cobalamin-dependent radical S-adenosylmethionine (SAM) methylase; however, TsrM does not reductively cleave SAM to the universal 5ʹ-deoxyadenosyl 5ʹ-radical intermediate, a hallmark of radical SAM (RS) enzymes. Herein, we report structures of TsrM from Kitasatospora setae, which are the first structures of a cobalamin-dependent radical SAM methylase. Unexpectedly, the structures show an essential arginine residue that resides in the proximal coordination sphere of the cobalamin cofactor, and a [4Fe–4S] cluster that is ligated by a glutamyl residue and three cysteines in a canonical CXXXCXXC RS motif. Structures in the presence of substrates suggest a substrate-assisted mechanism of catalysis, wherein the carboxylate group of SAM serves as a general base to deprotonate N1 of the tryptophan substrate, facilitating the formation of a C2 carbanion.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Atomic coordinates and structure factors for the reported crystal structures in this work have been deposited to the Protein Data Bank under accession numbers 6WTE (native KsTsrM) and 6WTF (aza-SAM- and Trp-bound KsTsrM). Source data are provided with this paper.
References
Kelly, W. L., Pan, L. & Li, C. X. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J. Am. Chem. Soc. 131, 4327–4334 (2009).
Bagley, M. C., Dale, J. W., Merritt, E. A. & Xiong, X. Thiopeptide antibiotics. Chem. Rev. 105, 685–714 (2005).
Millour, J. & Lam, E. W. FOXM1 is a transcriptional target of ERα and has a critical role in breast cancer endocrine sensitivity and resistance. Breast Cancer Res. 12, S3 (2010).
Hegde, N. S., Sanders, D. A., Rodriguez, R. & Balasubramanian, S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat. Chem. 3, 725–731 (2011).
Liao, R. J. et al. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 16, 141–147 (2009).
Frenzel, T., Zhou, P. & Floss, H. G. Formation of 2-methyltryptophan in the biosynthesis of thiostrepton: isolation of S-adenosylmethionine: tryptophan 2-methyltransferase. Arch. Biochem. Biophys. 278, 35–40 (1990).
Pierre, S. et al. Thiostrepton tryptophan methyltransferase expands the chemistry of radical SAM enzymes. Nat. Chem. Biol. 8, 957–959 (2012).
Holliday, G. L. et al. Atlas of the radical SAM superfamily: divergent evolution of function using a “plug and play” domain. Methods Enzymol. 606, 1–71 (2018).
Blaszczyk, A. J. et al. Spectroscopic and electrochemical characterization of the iron–sulfur and cobalamin cofactors of TsrM, an unusual radical S-adenosylmethionine methylase. J. Am. Chem. Soc. 138, 3416–3426 (2016).
Blaszczyk, A. J., Wang, B., Silakov, A., Ho, J. V. & Booker, S. J. Efficient methylation of C2 in l-tryptophan by the cobalamin-dependent radical S-adenosylmethionine methylase TsrM requires an unmodified N1 amine. J. Biol. Chem. 292, 15456–15467 (2017).
Bridwell-Rabb, J. & Drennan, C. L. Vitamin B12 in the spotlight again. Curr. Opin. Chem. Biol. 37, 63–70 (2017).
Bridwell-Rabb, J., Zhong, A. S., Sun, H. G., Drennan, C. L. & Liu, H. W. A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis. Nature 544, 322–326 (2017).
Drennan, C. L., Huang, S., Drummond, J. T., Matthews, R. G. & Ludwig, M. L. How a protein binds B12: a 3.0-angstrom X-ray structure of B12-binding domains of methionine synthase. Science 266, 1669–1674 (1994).
Kime, N. E. & Ibers, J. A. Co(III)–N bond-length in relation to Co(II)–N bond-length. The crystal structure of hexaamminecobalt(III) iodide [Co(NH3)6]I3. Acta Crystallograph. B 25, 168–169 (1969).
Kräutler, B. Thermodynamic trans-effects of the nucleotide base in the B12 coenzymes. Helv. Chim. Acta 70, 1268–1278 (1987).
Dowling, D. P. et al. Molecular basis of cobalamin-dependent RNA modification. Nucleic Acids Res. 44, 9965–9976 (2016).
Vey, J. L. et al. Structural basis for glycyl radical formation by pruvate formate-lyase activating enzyme. Proc. Natl Acad. Sci. USA 105, 16137–16141 (2008).
Broderick, J. B., Duffus, B. R., Duschene, K. S. & Shepard, E. M. Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 (2014).
Horitani, M. et al. Radical SAM catalysis via an organometallicintermediate with an Fe-[5′-C]-deoxyadenosyl bond. Science 352, 822–825 (2016).
Blaszczyk, A. J., Knox, H. L. & Booker, S. J. Understanding the role of electron donors in the reaction catalyzed by Tsrm, a cobalamin-dependent radical S-adenosylmethionine methylase. J. Biol. Inorg. Chem. 24, 831–839 (2019).
Hinckley, G. T., Ruzicka, F. J., Thompson, M. J., Blackburn, G. M. & Frey, P. A. Adenosyl coenzyme and pH dependence of the [4Fe–4S]2+/1+ transition in lysine 2,3-aminomutase. Arch. Biochem. Biophys. 414, 34–39 (2003).
Vey, J. L. & Drennan, C. L. Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 111, 2487–2506 (2011).
Cleland, W. W. in The Enzymes Vol. 2 (ed. Boyer, P. D.) 1–65 (Academic Press, 1970).
Lanz, N. D. et al. Enhanced solubilization of class B radical S-adenosylmethionine methylases by improved cobalamin uptake in Escherichia coli. Biochemistry 57, 1475–1490 (2018).
Wang, B. et al. Stereochemical and mechanistic investigation of the reaction catalyzed by Fom3 from Streptomyces fradiae, a cobalamin-dependent radical S-adenosylmethionine methylase. Biochemistry 57, 4972–4984 (2018).
Blaszczyk, A. J., Wang, R. X. & Booker, S. J. TsrM as a model for purifying and characterizing cobalamin-dependent radical S-adenosylmethionine methylases. Methods Enzymol. 595, 303–329 (2017).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Bunkoczi, G. et al. Phaser.MRage: automated molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 69, 2276–2286 (2013).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
PyMOL Molecular Graphics Systems, version 2.0 (Schrödinger).
Kung, Y., Doukov, T. I., Seravalli, J., Ragsdale, S. W. & Drennan, C. L. Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Biochemistry 48, 7432–7440 (2009).
Harding, M. M., Nowicki, M. W. & Walkinshaw, M. D. Metals in protein structures: a review of their principal features. Crystallogr. Rev. 16, 247–302 (2010).
Iwig, D. F. & Booker, S. J. Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-l-methionine. Biochemistry 43, 13496–13509 (2004).
Joce, C., Caryl, J., Stockley, P. G., Warriner, S. & Nelson, A. Identification of stable S-adenosylmethionine (SAM) analogues derivatised with bioorthogonal tags: effect of ligands on the affinity of the E. coli methionine repressor, MetJ, for its operator DNA. Org. Biomol. Chem. 7, 635–638 (2009).
Xiong, H. S., Stanley, B. A. & Pegg, A. E. Role of cysteine-82 in the catalytic mechanism of human S-adenosylmethionine decarboxylase. Biochemistry 38, 2462–2470 (1999).
Lanz, N. D. et al. RImN and AtsB as models for the overproduction and characterization radical SAM proteins. Methods Enzymol. 516, 125–152 (2012).
Olsson, M. H., Sondergaard, C. R., Rostkowski, M. & Jensen, J. H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011).
Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 (2013).
Friesner, R. A. et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein–ligand complexes. J. Med. Chem. 49, 6177–6196 (2006).
Friesner, R. A. et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004).
Halgren, T. A. et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759 (2004).
Acknowledgements
This work was supported by the NIH (GM-122595 to S.J.B. and GM-126982 to C.L.D.) and the Eberly Family Distinguished Chair in Science (S.J.B.). S.J.B. and C.L.D. are investigators of the Howard Hughes Medical Institute. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. Use of GM/CA@APS has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). This research also used the resources of the Berkeley Center for Structural Biology supported in part by the Howard Hughes Medical Institute. The Advanced Light Source is a DOE Office of Science User Facility under contract no. DE-AC02-05CH11231. The ALS-ENABLE beamlines are supported in part by the National Institutes of Health, National Institute of General Medical Sciences grant P30 GM124169.
Author information
Authors and Affiliations
Contributions
H.L.K. and S.J.B. developed the research plan and experimental strategy. H.L.K. isolated and crystallized proteins, and H.L.K. and E.L.S. collected the crystallographic data. H.L.K. and A.J.B. performed biochemical experiments. H.L.K., P.Y.-T.C., T.L.G., E.L.S., S.J.B. and C.L.D. analyzed and interpreted the crystallographic data. B.W. synthesized substrates and substrate analogs. A.M. performed computational docking experiments. H.L.K., P.Y.-T.C., S.J.B. and C.L.D. wrote the manuscript, while all other authors reviewed and commented on it.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Topology diagram of KsTsrM depicting the different domains.
The blue dot represents the conserved Arg residue that occupies the lower axial position of the cobalamin, which is shown as two purple lines. The three yellow spheres on the loop after α1 of the TIM barrel represent the three conserved cysteines that coordinate the [4Fe-4S] cluster. The orange sphere on the loop following β2 of the TIM barrel is a conserved Glu residue that coordinates the unique Fe of the cluster.
Extended Data Fig. 2 Structural comparison of TsrM and OxsB.
Ribbon diagrams of (a) KsTsrM and (b) OxsB with domains colored to highlight structural similarities: Rossman fold (teal), TIM barrel (light blue), and C-terminal domain (purple). The N-terminal domain of OxsB is colored pink. Overlays of the Rossmann fold (c), TIM barrel (d) and C-terminal domain (e) of KsTsrM (teal, light blue, and purple respectively) with OxsB (grey).
Extended Data Fig. 3 Binding of cobalamin in the Rossmann fold of TsrM.
Conserved stabilizing residues among TsrMs are depicted.
Extended Data Fig. 4 Comparison of lower axial ligands of cobalamin.
The different axial ligands in (a) KsTsrM, (b) OxsB, and (c) MetH.
Extended Data Fig. 5 Time-dependent production of MeTrp by wild-type KsTsrM and the Arg69Lys variant.
a, Time-dependent production of MeTrp by KsTsrM (1 µM) using the flavodoxin/flavodoxin reductase/NADPH reducing system. kcat is 14 min−1. b, UV-vis analysis of KsTsrM. KsTsrM was reduced with Ti (III) citrate to form cob(I)alamin. Upon addition of SAM to the reaction, the peak at 390 nm, indicative of cob(I)alamin, decays, while a peak at 520 nm, indicative of MeCbl, grows in. c, Time-dependent production of MeTrp by the KsTsrM R69K variant using either Ti (III) citrate (red) or Fld/Fld Red/NADPH (purple) as reductant. Despite using 75 µM enzyme (indicated by dotted line), not even a full turnover is observed over 20 min by KsTsrM R69K with either reducing system. The inset displays a reduced Y-axis range to allow the small amount of activity in the presence of Ti (III) citrate to be observed. d, UV-vis analysis of KsTsrM R69K. As in (b), KsTsrM R69K was reduced with Ti (III) citrate to generate cob(I)alamin. When SAM is added, cob(I)alamin decays and MeCbl grows in, indicating that MeCbl formation is not significantly diminished with this variant. Error bars represent the standard deviation of reactions conducted in triplicate, with the center point representing the average.
Extended Data Fig. 6 Different roles of Arg residues that occupy the proximal face of cobalamin.
Comparison of the lower proximal face of cobalamin in (a) KsTsrM and (b) QueG (PDB: 5D08).
Extended Data Fig. 7 Computational docking studies of Trp positioning in active site.
a, Overlay of the two most likely poses for Trp in the computational docking model: pose 1 (lavender) and pose 2 (pink). b, Comparison of computational docking model pose 1 (lavender) to flipped Trp observed in crystal structure (teal). c, Associated distances between carboxylate of aza-SAM to Trp in docking model pose 2. d, Alternative view of pose 2, demonstrating distances from aza-SAM.
Extended Data Fig. 8 Comparison of Tyr308 in native and substrate-bound structures.
a, Overlay of KsTsrM native structure (teal) with substrate bound structure (light blue), showing change in conformation of Tyr308. b, Positioning of Tyr308 in docking model with pose 2, showing proximity to C2 position of Trp.
Extended Data Fig. 9 Analysis of MeCbl formation using dcSAM.
a, LC-MS/MS analysis of MeCbl formation in a reaction containing TsrM (10 µM), flavodoxin/flavodoxin reductase/NADPH, and dcSAM (1.5 mM). b, Time-dependent formation of MeCbl. Error bars represent the standard deviation of triplicate determinations. The dashed line indicates the concentration of enzyme in the reaction, with the central point representing the average.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and Table 1.
Source data
Source Data Fig. 4
Raw data for Fig. 4d,e.
Source Data Extended Data Fig. 5
Raw data for Extended Data Fig. 5a,c.
Source Data Extended Data Fig. 9
Raw data for Extended Data Fig. 9a,b.
Rights and permissions
About this article
Cite this article
Knox, H.L., Chen, P.YT., Blaszczyk, A.J. et al. Structural basis for non-radical catalysis by TsrM, a radical SAM methylase. Nat Chem Biol 17, 485–491 (2021). https://doi.org/10.1038/s41589-020-00717-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-00717-y
This article is cited by
-
A modular and synthetic biosynthesis platform for de novo production of diverse halogenated tryptophan-derived molecules
Nature Communications (2024)
-
Crystallographic snapshots of a B12-dependent radical SAM methyltransferase
Nature (2022)
-
Structure of a B12-dependent radical SAM enzyme in carbapenem biosynthesis
Nature (2022)