Abstract
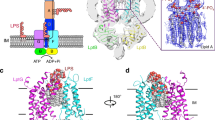
Lipopolysaccharide (LPS) transport to the outer membrane (OM) is a crucial step in the biogenesis of microbial surface defenses. Although many features of the translocation mechanism have been elucidated, molecular details of LPS insertion via the LPS transport (Lpt) OM protein LptDE remain elusive. Here, we integrate native MS with hydrogen–deuterium exchange MS and molecular dynamics simulations to investigate the influence of substrate and peptide binding on the conformational dynamics of LptDE. Our data reveal that LPS induces opening of the LptD β-taco domain, coupled with conformational changes on β-strands adjacent to the putative lateral exit gate. Conversely, an antimicrobial peptide, thanatin, stabilizes the β-taco, thereby preventing LPS transport. Our results illustrate that LPS insertion into the OM relies on concerted opening movements of both the β-barrel and β-taco domains of LptD, and suggest a means for developing antimicrobial therapeutics targeting this essential process in Gram-negative ESKAPE pathogens.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data supporting the findings of this study are available from the corresponding authors upon reasonable request. HDX-MS raw data and the HDX data tables have been deposited to the ProteomeXchange Consortium via the PRIDE60 partner repository with the dataset identifier PXD021743.
The structural models employed in this study are accessible through the PDB (https://www.rcsb.org/) under accession nos. 5IV9 (KpLptDE), 5XO4 (thanatin), 2R1A (EcLptA), 6GD5 (EcLptA–thanatin complex) and 4Q35 (SfLptDE). Source data are provided with this paper.
References
Whitfield, C. & Trent, M. S. Biosynthesis and export of bacterial lipopolysaccharides. Ann. Rev. Biochem. 83, 99–128 (2014).
Okuda, S., Sherman, D. J., Silhavy, T. J., Ruiz, N. & Kahne, D. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 14, 337–345 (2016).
Ruiz, N., Kahne, D. & Silhavy, T. J. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat. Rev. Microbiol. 7, 677–683 (2009).
Luo, Q. et al. Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat. Struct. Mol. Biol. 24, 469–474 (2017).
Li, Y., Orlando, B. J. & Liao, M. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567, 486–490 (2019).
Tang, X. et al. Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism. Nat. Commun. 10, 4175 (2019).
Owens, T. W. et al. Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553 (2019).
Villa, R. et al. The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J. Bacteriol. 195, 1100–1108 (2013).
Chng, S. S., Ruiz, N., Chimalakonda, G., Silhavy, T. J. & Kahne, D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl Acad. Sci. USA 107, 5363–5368 (2010).
Freinkman, E., Chng, S. S. & Kahne, D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl Acad. Sci. USA 108, 2486–2491 (2011).
Qiao, S., Luo, Q., Zhao, Y., Zhang, X. C. & Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014).
Dong, H. et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56 (2014).
Botos, I. et al. Structural and functional characterization of the LPS transporter LptDE from Gram-negative pathogens. Structure 24, 965–976 (2016).
Li, X., Gu, Y., Dong, H., Wang, W. & Dong, C. Trapped lipopolysaccharide and LptD intermediates reveal lipopolysaccharide translocation steps across the Escherichia coli outer membrane. Sci. Rep. 5, 11883 (2015).
Gu, Y. et al. Lipopolysaccharide is inserted into the outer membrane through an intramembrane hole, a lumen gate, and the lateral opening of LptD. Structure 23, 496–504 (2015).
World Health Organization. Global Priority List of Antibiotic-resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics (WHO, 2017); http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/
Srinivas, N. et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013 (2010).
Andolina, G. et al. A peptidomimetic antibiotic interacts with the periplasmic domain of LptD from Pseudomonas aeruginosa. ACS Chem. Biol. 13, 666–675 (2018).
Werneburg, M. et al. Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. ChemBioChem 13, 1767–1775 (2012).
Fehlbaum, P. et al. Structure–activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc. Natl Acad. Sci. USA 93, 1221–1225 (1996).
Vetterli, S. U. et al. Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci. Adv. 4, eaau2634 (2018).
Bolla, J. R. et al. Direct observation of the influence of cardiolipin and antibiotics on lipid II binding to MurJ. Nat. Chem. 10, 363–371 (2018).
Fiorentino, F., Bolla, J. R., Mehmood, S. & Robinson, C. V. The different effects of substrates and nucleotides on the complex formation of ABC transporters. Structure 27, 651–659 (2019).
Yen, H. Y. et al. PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 559, 423–427 (2018).
Bolla, J. R., Howes, A. C., Fiorentino, F. & Robinson, C. V. Assembly and regulation of the chlorhexidine-specific efflux pump AceI. Proc. Natl Acad. Sci. USA 117, 17011–17018 (2020).
Martens, C. et al. Direct protein–lipid interactions shape the conformational landscape of secondary transporters. Nat. Commun. 9, 4151 (2018).
Nielsen, A. K. et al. Substrate-induced conformational dynamics of the dopamine transporter. Nat. Commun. 10, 2714 (2019).
Landreh, M. et al. Integrating mass spectrometry with MD simulations reveals the role of lipids in Na+/H+ antiporters. Nat. Commun. 8, 13993 (2017).
Skinner, J. J. et al. Benchmarking all-atom simulations using hydrogen exchange. Proc. Natl Acad. Sci. USA 111, 15975–15980 (2014).
Persson, F. & Halle, B. How amide hydrogens exchange in native proteins. Proc. Natl Acad. Sci. USA 112, 10383–10388 (2015).
Laganowsky, A., Reading, E., Hopper, J. T. & Robinson, C. V. Mass spectrometry of intact membrane protein complexes. Nat. Protoc. 8, 639–651 (2013).
Raetz, C. R. et al. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 47, 1097–1111 (2006).
Xie, R., Taylor, R. J. & Kahne, D. Outer membrane translocon communicates with inner membrane ATPase to stop lipopolysaccharide transport. J. Am. Chem. Soc. 140, 12691–12694 (2018).
Bolla, J. R. et al. A mass-spectrometry-based approach to distinguish annular and specific lipid binding to membrane proteins. Angew. Chem. Int. Ed. 59, 3523–3528 (2020).
Masson, G. R. et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 16, 595–602 (2019).
Lundquist, K. P. & Gumbart, J. C. Presence of substrate aids lateral gate separation in LptD. Biochim. Biophys. Acta Biomembr. 1862, 183025 (2020).
Merkle, P. S. et al. Substrate-modulated unwinding of transmembrane helices in the NSS transporter LeuT. Sci. Adv. 4, eaar6179 (2018).
Zhou, J. et al. Conformational dynamics of 1-deoxy-d-xylulose 5-phosphate synthase on ligand binding revealed by H/D exchange MS. Proc. Natl Acad. Sci. USA 114, 9355–9360 (2017).
Jefferys, E., Sands, Z. A., Shi, J., Sansom, M. S. P. & Fowler, P. W. Alchembed: a computational method for incorporating multiple proteins into complex lipid geometries. J. Chem. Theory Comput. 11, 2743–2754 (2015).
Bonomi, M. et al. PLUMED: a portable plugin for free-energy calculations with molecular dynamics. Comput. Phys. Commun. 180, 1961–1972 (2009).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Tomasek, D. et al. Structure of a nascent membrane protein as it folds on the BAM complex. Nature 583, 473–478 (2020).
Hernandez, H. & Robinson, C. V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2, 715–726 (2007).
Marty, M. T. et al. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 (2015).
Cubrilovic, D. et al. Determination of protein–ligand binding constants of a cooperatively regulated tetrameric enzyme using electrospray mass spectrometry. ACS Chem. Biol. 9, 218–226 (2014).
Guttman, M. et al. Tuning a high transmission ion guide to prevent gas-phase proton exchange during H/D exchange MS analysis. J. Am. Soc. Mass Spectrom. 27, 662–668 (2016).
Lau, A. M. C., Ahdash, Z., Martens, C. & Politis, A. Deuteros: software for rapid analysis and visualization of data from differential hydrogen deuterium exchange-mass spectrometry. Bioinformatics 35, 3171–3173 (2019).
Weis, D. D., Engen, J. R. & Kass, I. J. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass Spectrom. 17, 1700–1703 (2006).
Guttman, M., Weis, D. D., Engen, J. R. & Lee, K. K. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J. Am. Soc. Mass Spectrom. 24, 1906–1912 (2013).
Konermann, L., Pan, J. & Liu, Y. H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Wiederstein, M. & Sippl, M. J. TopMatch-web: pairwise matching of large assemblies of protein and nucleic acid chains in 3D. Nucleic Acids Res. 48, W31–W35 (2020).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Monticelli, L. et al. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 4, 819–834 (2008).
Wassenaar, T. A., Ingólfsson, H. I., Böckmann, R. A., Tieleman, D. P. & Marrink, S. J. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 11, 2144–2155 (2015).
Stansfeld, P. J. & Sansom, M. S. P. From coarse grained to atomistic: a serial multiscale approach to membrane protein simulations. J. Chem. Theory Comput. 7, 1157–1166 (2011).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2020).
Acknowledgements
Research in C.V.R.’s laboratory is supported by a Medical Research Council program grant (MR/N020413/1), a European Research Council Advanced Grant ENABLE (695511) and a Wellcome Trust Senior Investigator Award (104633/Z/14/Z). Research in P.J.S.’s laboratory is funded by Wellcome (208361/Z/17/Z), the MRC (MR/S009213/1) and BBSRC (BB/P01948X/1, BB/R002517/1 and BB/S003339/1). This project made use of time on ARCHER and JADE, granted via the UK High-End Computing Consortium for Biomolecular Simulation (HECBioSim). P.J.S. acknowledges Athena at HPC Midlands+, funded by the EPSRC under grant no. EP/P020232/1 and the University of Warwick Scientific Computing Research Technology Platform for computational access. F.F. holds a SABS CDT studentship supported by the EPSRC and MRC (EP/L016044/1). J.B.S. is supported by the Oxford interdisciplinary DTP and the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/M011224/1). We also thank S. Roy (University of Oxford) for his help with HDX-MS data analysis and rendering.
Author information
Authors and Affiliations
Contributions
F.F., J.R.B., P.J.S. and C.V.R. designed the research. F.F. expressed and purified the protein samples and performed all nMS measurements. F.F. and J.R.B. analyzed nMS data. F.F. and X.Q. collected HDX-MS data. F.F. analyzed and interpreted HDX-MS data with the help of X.Q., J.R.B. and S.M. J.B.S. performed the MD simulations with the assistance of P.J.S., who modeled the initial substrate-bound LptDE states with B.M.-W. J.B.S. analyzed MD simulations data with the help of R.A.C., C.K.C. and P.J.S. F.F., J.B.S., R.A.C., C.K.C., J.R.B., P.J.S. and C.V.R. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.V.R. is a co-founder and consultant at OMass Therapeutics. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Detergent competition experiments to assess LptDE lipid binding.
LptDE (5 μM, left) in 0.5% (w/v) C8E4 was mixed with 10 μM Re-LPS (a, orange adducts), POPG (b, red adducts) or CDL (c, magenta adducts). The protein-lipid mixture was then supplemented with increasing concentrations of n-nonyl-β-D-glucopyranoside (NG). Detergent addition decreases POPG and CDL binding, but has no pronounced effect on Re-LPS.
Extended Data Fig. 2 Thanatin binding efficiency and effect on Re-LPS binding.
a, Mass spectra recorded for solutions of LptDE (5 μM) with increasing concentrations of thanatin (green adducts). b, Mass spectra of Re-LPS-bound LptDE supplemented with 1 μM thanatin (as described in Fig. 1d) with a focus on the 16+ charge state showing the presence of the 2:1 Re-LPS:thanatin complex, particularly at higher Re-LPS concentration. c, Quantification of the total amount of Re-LPS bound to LptDE in the absence or in the presence of thanatin (related to Fig. 1d). Error bars represent s.d. (n = 3). d, Thanatin-first nMS analysis: LptDE (5 μM) was initially mixed with thanatin and then supplemented with Re-LPS (10 μM).
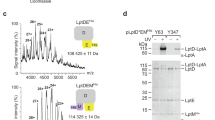
Extended Data Fig. 3 Sequence coverage of LptDE.
68 peptides covering 70.5% of LptD sequence a, and 17 peptides covering 87.4% of LptE b, were identified following digestion with immobilized pepsin.
Extended Data Fig. 4 Representative mass spectra for peptides showing EX1/EXX kinetics.
Mass spectra are shown for apo-LptDE, LPS, thanatin or LPS + thanatin states. Two binomial isotopic envelopes produced the best fit for the spectra yielding low- (green) and high-mass (light blue) populations. The sums of the two binomial distributions are shown in red. a, Peptide 66-92. b, Peptide 105-115. c, Peptide 116-19. d, Peptide 119-129. e, Peptide 130-141. f, Peptide 171-179.
Extended Data Fig. 5 Monoexponential fitting of the high-mass population in peptides showing EX1 kinetics.
Extracted relative abundances of high- mass populations plotted as a function of labeling time and fitted to a single exponential function with a variable intercept and plateau (to account for the lack of saturation seen in most peptides) to obtain the rate of translation from the low-mass population to the high-mass population (kop) and the half-life of the low mass population (t1/2). The apo state is indicated in purple, LPS-bound state in orange, thanatin-bound state in green, and LPS + thanatin state in maroon. Standard deviations are plotted as error bars (nbiological = 2; ntechnical = 3) but are in some instances too small to be visible. In the case of thanatin-bound state and LPS + thanatin state kinetic values could not be extracted because of poor fitting (the increase of high-mass population is incremental given the slow kinetics). In these cases, the dotted line is included only for visual guidance.
Extended Data Fig. 6 Re-LPS contacts and position within the β-taco.
a, Average percentage occupancy of Re-LPS contacts made with the β-taco with a 4 Å cut-off. These contacts have been compared to those made by the detergent in the SfLptD structure (PDB ID: 4Q35; marked by an asterisk) and found that 90% of residues which interact with detergent. b, Top: Kernel density estimate (KDE) of the center of mass (COM) position between the five replicates indicate minimal diffusion over the course of the simulation. Middle: Partial density of Re-LPS across the β-taco demonstrating consistent and rigid contact across the domain, this implies that the Re-LPS bound within the β-taco does not have enough simulation time to sample the entire soluble domain in different binding poses, which would account for the discrepancy in deprotected coverage across this domain when compared to HDX-MS (Supplementary Fig. 15). Bottom: reference cartoon structure of β-taco to compare with above plot axes.
Extended Data Fig. 7 Re-LPS contact mapped onto LptDE.
a, Average Re-LPS contacts mapped onto cartoon representations of LptDE. b, Residues interacting with the three Re-LPS lipids referenced in Fig. 2c (Re-LPS(1-3)) are shown in stick representation. c, C-terminal strand of the β-taco is in contact with the bilayer (Supplementary Fig. 15c). d, Ile186 and Phe187 are in transient contact with the Re-LPS lipids engaged with the lateral putative exit gate of the β-barrel. The open gate causes Re-LPS lipids (shown in yellow and pink) to be laterally pulled into the β-barrel sinking them into the inner leaflet of the and allowing the tails to interact with the C-terminal strand of the β-taco. This may suggest how Re-LPS gets laterally extruded into the OM during the translocation process.
Extended Data Fig. 8 Re-LPS distance plots.
Top: snapshot of the key lipid interactions between Re-LPS and the lateral gate peptide (232-251). Bottom: Plots measuring the distance from the geometric center of the peptide (232-251) to the geometric center of the lipids. Data plotted as the average of five repeats throughout the course of the simulation, standard deviation shaded gray.
Extended Data Fig. 9 Sequence and structure alignments of KpLptD and EcLptA.
a, Sequence alignment between KpLptD and EcLptA. Red color indicates conserved residues, yellow indicates highly similar residues. Circles indicate crucial residues for LptA-thanatin interaction. Blue color represents van-der-Waals interactions while green represents ionic bridges and H-bonds. b, Structure alignment of LptD (PDB ID: 5IV9) with apo-LptA (PDB ID: 2R1A). The RMSD is equal to 1.48 Å over 115 Cα.
Extended Data Fig. 10 Evaluation of LptD-thanatin interaction stability.
a, RMSD of thanatin docked to the bottom (i) of the β-jellyroll in its closed conformation compared to being docked to the top (ii) where standard deviation is colored gray. b, Right: schematic showing the orientation of the β-taco vector drawn through Cα of residues Asp33 and Pro190, where x, y displacement is shown as red arrows. Right: x,y displacement of the β-taco vector mapped relative to the origin 0.0, 0.0 (defined as the original position of β-taco in the crystal structure) comparing the conformation space sampled of the apo-β-taco against when thanatin is docked to the (i) bottom and (ii) top of the β-taco.
Supplementary information
Supplementary Information
Supplementary Figs. 1–18 and Tables 1–5.
Supplementary Video
Morph of LptDE complex. Morph between the closed conformation of the complex and the open conformation of the complex, with disulfide bonds between cysteines at the β-taco (Cys7, Cys149) and the β-barrel (Cys696, Cys697) shown as spheres.
Source data
Source Data Fig. 2
HDX-MS data used to create deuterium uptake plots for apo and LPS-bound protein and to map the difference in relative deuterium uptake (LPS - apo) onto LptD structure.
Source Data Figs. 3 and 4
HDX-MS data used to create deuterium uptake plots for apo, thantin-bound and LPS + thanatin bound protein and to map the difference in relative deuterium uptake (ligand bound - apo) onto LptD structure.
Rights and permissions
About this article
Cite this article
Fiorentino, F., Sauer, J.B., Qiu, X. et al. Dynamics of an LPS translocon induced by substrate and an antimicrobial peptide. Nat Chem Biol 17, 187–195 (2021). https://doi.org/10.1038/s41589-020-00694-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-00694-2
This article is cited by
-
LptM promotes oxidative maturation of the lipopolysaccharide translocon by substrate binding mimicry
Nature Communications (2023)
-
TPGS-based and S-thanatin functionalized nanorods for overcoming drug resistance in Klebsiella pneumonia
Nature Communications (2022)
-
Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation
Nature Communications (2022)
-
NMR Structure and Localization of the Host Defense Peptide ThanatinM21F in Zwitterionic Dodecylphosphocholine Micelle: Implications in Antimicrobial and Hemolytic Activity
The Journal of Membrane Biology (2022)
-
Antimicrobial peptides: mechanism of action, activity and clinical potential
Military Medical Research (2021)