Abstract
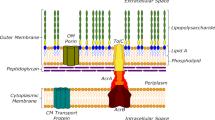
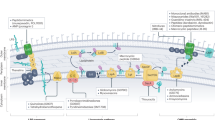
We live in the era of antibiotic resistance, and this problem will progressively worsen if no new solutions emerge. In particular, Gram-negative pathogens present both biological and chemical challenges that hinder the discovery of new antibacterial drugs. First, these bacteria are protected from a variety of structurally diverse drugs by a low-permeability barrier composed of two membranes with distinct permeability properties, in addition to active drug efflux, making this cell envelope impermeable to most compounds. Second, chemical libraries currently used in drug discovery contain few compounds that can penetrate Gram-negative bacteria. As a result of these challenges, intensive screening campaigns have led to few successes, highlighting the need for new approaches to identify regions of chemical space that are specifically relevant to antibacterial drug discovery. Herein we provide an overview of emerging insights into this problem and outline a general approach to addressing it using prospective analysis of chemical libraries for the ability of compounds to accumulate in Gram-negative bacteria. The overall goal is to develop robust cheminformatic tools to predict Gram-negative permeation and efflux, which can then be used to guide medicinal chemistry campaigns and the design of antibacterial discovery libraries.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Antibiotic Resistance Threats in the United States 2019 (Centers for Disease Control and Prevention, 2019); https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf
Antimicrobial Resistance: Global Report on Surveillance 2014 (World Health Organization, 2014); http://www.who.int/drugresistance/documents/surveillancereport/en/
A Scientific Roadmap for Antibiotic Discovery (The Pew Charitable Trusts, 2016); https://www.pewtrusts.org/en/research-and-analysis/reports/2016/05/a-scientific-roadmap-for-antibiotic-discovery
Rice, L. B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081 (2008).
De Oliveira, D. M. P. et al. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33, e00181 (2020).
Tracking the Global Pipeline of Antibiotics in Development, April 2020 (The Pew Charitable Trusts, 2020); https://www.pewtrusts.org/en/research-and-analysis/issue-briefs/2020/04/tracking-the-global-pipeline-of-antibiotics-in-development
Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003).
Zgurskaya, H. I., Lopez, C. A. & Gnanakaran, S. Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect. Dis. 1, 512–522 (2015).
Silver, L. L. A Gestalt approach to Gram-negative entry. Bioorg. Med. Chem. 24, 6379–6389 (2016).
Masi, M., Refregiers, M., Pos, K. M. & Pages, J.-M. Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat. Microbiol. 2, 17001 (2017).
Tommasi, R., Iyer, R. & Miller, A. A. Antibacterial drug discovery: some assembly required. ACS Infect. Dis. 4, 686–695 (2018).
Zgurskaya, H. I. & Rybenkov, V. V. Permeability barriers of Gram-negative pathogens. Ann. N. Y. Acad. Sci. 1459, 5–18 (2020).
Saha, P., Sikdar, S., Krishnamoorthy, G., Zgurskaya, H. I. & Rybenkov, V. V. Drug permeation against efflux by two transporters. ACS Infect. Dis. 6, 747–758 (2020).
Westfall, D. A. et al. Bifurcation kinetics of drug uptake by Gram-negative bacteria. PLoS ONE 12, e0184671 (2017). A kinetic model that accurately describes small-molecule permeation of the Gram-negative envelope was developed. The model-predicted complex, nonlinear patterns dependent on concentration and time that were validated experimentally.
Krishnamoorthy, G. et al. Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. mBio 8, e01172 (2017). Analysis of the activity of antibiotics in four-strain sets of wild-type, hyperporinated, efflux-deficient, and doubly-compromised A. baumannii, P. aeruginosa, and Burkholderia spp. revealed a synergistic relationship between efflux and the permeability barrier in protecting these bacteria from structurally diverse antibiotics.
Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discovery 6, 29–40 (2007).
Tommasi, R., Brown, D. G., Walkup, G. K., Manchester, J. I. & Miller, A. A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discovery 14, 529–542 (2015).
Lewis, K. Antibiotics: Recover the lost art of drug discovery. Nature 485, 439–440 (2012).
Kamio, Y. & Nikaido, H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570 (1976).
Raetz, C. R. & Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 (2002).
Henderson, J. C. et al. The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu. Rev. Microbiol. 70, 255–278 (2016).
Vergalli, J. et al. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 18, 164–176 (2020).
Zgurskaya, H. I., Rybenkov, V. V., Krishnamoorthy, G. & Leus, I. V. Trans-envelope multidrug efflux pumps of Gram-negative bacteria and their synergism with the outer membrane barrier. Res. Microbiol. 169, 351–356 (2018).
Poirel, L., Jayol, A. & Nordmann, P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596 (2017).
Pagnout, C. et al. Pleiotropic effects of rfa-gene mutations on Escherichia coli envelope properties. Sci. Rep. 9, 9696 (2019).
Krishnamoorthy, G. et al. Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob. Agents Chemother. 60, 7372–7381 (2016).
Leus, I. V. et al. Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J. Bacteriol. 200, e00049 (2018).
Krishnamoorthy, G. et al. Efflux pumps of Burkholderia thailandensis control the permeability barrier of the outer membrane. Antimicrob. Agents Chemother. 63, e00956 (2019).
Sohlenkamp, C. & Geiger, O. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159 (2016).
Richter, M. F. & Hergenrother, P. J. The challenge of converting Gram-positive-only compounds into broad-spectrum antibiotics. Ann. N. Y. Acad. Sci. 1435, 18–38 (2019).
Richter, M. F. et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 545, 299–304 (2017). Screening of a library of 100 diverse, natural-product-derived compounds for compound accumulation in E. coli identified a primary amine, amphiphilic moment, low globularity, and rigidity as factors associated with accumulation, enabling design of a Gram-negative-active analogue of a Gram-positive-restricted antibiotic.
Parker, E. N. et al. Implementation of permeation rules leads to a FabI inhibitor with activity against Gram-negative pathogens. Nat. Microbiol. 5, 67–75 (2020).
Motika, S. E. et al. Gram-negative antibiotic active through inhibition of an essential riboswitch. J. Am. Chem. Soc. 142, 10856–10862 (2020).
Isabella, V. M. et al. Toward the rational design of carbapenem uptake in Pseudomonas aeruginosa. Chem. Biol. 22, 535–547 (2015).
Li, Y. et al. First-generation structure-activity relationship studies of 2,3,4,9-tetrahydro-1H-carbazol-1-amines as CpxA phosphatase inhibitors. Bioorg. Med. Chem. Lett. 29, 1836–1841 (2019).
Green, A. T. et al. Discovery of multidrug efflux pump inhibitors with a novel chemical scaffold. Biochim. Biophys. Acta Gen. Subj. 1864, 129546 (2020).
Nestorovich, E. M., Danelon, C., Winterhalter, M. & Bezrukov, S. M. Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc. Natl Acad. Sci. USA 99, 9789–9794 (2002).
Wang, J., Terrasse, R., Bafna, J. A., Benier, L. & Winterhalter, M. Electrophysiological characterization of transport across outer-membrane channels from Gram-negative bacteria in presence of lipopolysaccharides. Angew. Chem. Int. Ed. 59, 8517–8521 (2020).
Acosta-Gutierrez, S. et al. Getting drugs into Gram-negative bacteria: rational rules for permeation through general porins. ACS Infect. Dis. 4, 1487–1498 (2018).
Cooper, S. J. et al. Molecular properties that define the activities of antibiotics in Escherichia coli and Pseudomonas aeruginosa. ACS Infect. Dis. 4, 1223–1234 (2018).
Iyer, R. et al. Whole-cell-based assay to evaluate structure permeation relationships for carbapenem passage through the Pseudomonas aeruginosa porin OprD. ACS Infect. Dis. 3, 310–319 (2017).
Nikaido, H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264, 382–388 (1994).
Iyer, R. et al. Evaluating LC-MS/MS to measure accumulation of compounds within bacteria. ACS Infect. Dis. 4, 1336–1345 (2018). Screening of a library of >100 DNA ligase inhibitors for compound accumulation in E. coli showed poor correlation between overall bacteria-associated compound levels and antibacterial activity in compounds with matched biochemical activities, highlighting the importance of subcellular localization.
Widya, M. et al. Development and optimization of a higher-throughput bacterial compound accumulation assay. ACS Infect. Dis. 5, 394–405 (2019).
O’Shea, R. & Moser, H. E. Physicochemical properties of antibacterial compounds: implications for drug discovery. J. Med. Chem. 51, 2871–2878 (2008).
Davis, T. D., Gerry, C. J. & Tan, D. S. General platform for systematic quantitative evaluation of small-molecule permeability in bacteria. ACS Chem. Biol. 9, 2535–2544 (2014). A platform for prospective, activity-independent analysis of compound accumulation in bacteria was developed. Analysis of a panel of acyl sulfamoyladenosines identified physicochemical properties that correlate with accumulation, enabling the design of analogues with increased accumulation.
Brown, D. G., May-Dracka, T. L., Gagnon, M. M. & Tommasi, R. Trends and exceptions of physical properties on antibacterial activity for Gram-positive and Gram-negative pathogens. J. Med. Chem. 57, 10144–10161 (2014).
Novick, R. P. Microiodometric assay for penicillinase. Biochem. J. 83, 236–240 (1962).
Zimmermann, W. & Rosselet, A. Function of the outer membrane of Escherichia coli as a permeability barrier to beta-lactam antibiotics. Antimicrob. Agents Chemother. 12, 368–372 (1977).
Murakami, K. & Yoshida, T. Penetration of cephalosporins and corresponding 1-oxacephalosporins through the outer layer of Gram-negative bacteria and its contribution to antibacterial activity. Antimicrob. Agents Chemother. 21, 254–258 (1982).
Nikaido, H., Rosenberg, E. Y. & Foulds, J. Porin channels in Escherichia coli: studies with beta-lactams in intact cells. J. Bacteriol. 153, 232–240 (1983).
Yoshimura, F. & Nikaido, H. Diffusion of β-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob. Agents Chemother. 27, 84–92 (1985).
Chopra, I. & Hacker, K. Uptake of minocycline by Escherichia coli. J. Antimicrob. Chemother. 29, 19–25 (1992).
Diver, J. M., Piddock, L. J. V. & Wise, R. The accumulation of five quinolone antibacterial agents by Escherichia coli. J. Antimicrob. Chemother. 25, 319–333 (1990).
Mortimer, P. G. S. & Piddock, L. J. V. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 28, 639–653 (1991).
Bazile, S., Moreau, N., Bouzard, D. & Essiz, M. Relationship among antibacterial activity, inhibition of DNA gyrase, and intracellular accumulation of 11 fluoroquinolones. Antimicrob. Agents Chemother. 36, 2622–2627 (1992). This is a seminal early study demonstrating the integration of compound accumulation levels with biochemical inhibitory activity to predict antibacterial activity in E. coli and P. aeruginosa.
McCaffrey, C., Bertasso, A., Pace, J. & Georgopapadakou, N. H. Quinolone accumulation in Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus. Antimicrob. Agents Chemother. 36, 1601–1605 (1992).
Asuquo, A. E. & Piddock, L. J. Accumulation and killing kinetics of fifteen quinolones for Escherichia coli, Staphylococcus aureus and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 31, 865–880 (1993).
Piddock, L. J. V., Jin, Y. F. & Griggs, D. J. Effect of hydrophobicity and molecular mass on the accumulation of fluoroquinolones by Staphylococcus aureus. J. Antimicrob. Chemother. 47, 261–270 (2001).
Mortimer, P. G. S. & Piddock, L. J. V. The accumulation of five antibacterial agents in porin-deficient mutants of Escherichia coli. J. Antimicrob. Chemother. 32, 195–213 (1993).
Piddock, L. J. V., Jin, Y. F., Ricci, V. & Asuquo, A. E. Quinolone accumulation by Pseudomonas aeruginosa, Staphylococcus aureus and Escherichia coli. J. Antimicrob. Chemother. 43, 61–70 (1999).
Cai, H., Rose, K., Liang, L.-H., Dunham, S. & Stover, C. Development of a liquid chromatography/mass spectrometry-based drug accumulation assay in Pseudomonas aeruginosa. Anal. Biochem. 385, 321–325 (2009).
Ferreras, J. A., Ryu, J.-S., Di Lello, F., Tan, D. S. & Quadri, L. E. N. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat. Chem. Biol. 1, 29–32 (2005).
Huigens, R. W. et al. A ring-distortion strategy to construct stereochemically complex and structurally diverse compounds from natural products. Nat. Chem. 5, 195–202 (2013).
O’Connor, C. J., Beckmann, H. S. G. & Spring, D. R. Diversity-oriented synthesis: producing chemical tools for dissecting biology. Chem. Soc. Rev. 41, 4444–4456 (2012).
Gerry, C. J. & Schreiber, S. L. Recent achievements and current trajectories of diversity-oriented synthesis. Curr. Opin. Chem. Biol. 56, 1–9 (2020).
Prochnow, H. et al. Subcellular quantification of uptake in Gram-negative bacteria. Anal. Chem. 91, 1863–1872 (2019).
Zhou, Y. et al. Thinking outside the “bug”: a unique assay to measure intracellular drug penetration in Gram-negative bacteria. Anal. Chem. 87, 3579–3584 (2015).
Blair, J. M. A. & Piddock, L. J. V. How to measure export via bacterial multidrug resistance efflux pumps. mBio 7, e00840 (2016).
Stone, M. R. L., Butler, M. S., Phetsang, W., Cooper, M. A. & Blaskovich, M. A. T. Fluorescent antibiotics: new research tools to fight antibiotic resistance. Trends Biotechnol. 36, 523–536 (2018).
Tanner, L., Denti, P., Wiesner, L. & Warner, D. F. Drug permeation and metabolism in Mycobacterium tuberculosis: prioritising local exposure as essential criterion in new TB drug development. IUBMB Life 70, 926–937 (2018).
Kaščáková, S., Maigre, L., Chevalier, J., Réfrégiers, M. & Pagès, J.-M. Antibiotic transport in resistant bacteria: synchrotron UV fluorescence microscopy to determine antibiotic accumulation with single cell resolution. PLoS ONE 7, e38624 (2012).
Vergalli, J. et al. Spectrofluorimetric quantification of antibiotic drug concentration in bacterial cells for the characterization of translocation across bacterial membranes. Nat. Protoc. 13, 1348–1361 (2018).
Paixão, L. et al. Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 3, 18 (2009).
Whittle, E. E. et al. Flow cytometric analysis of efflux by dye accumulation. Front. Microbiol. 10, 2319 (2019).
Prideaux, B. et al. High-sensitivity MALDI-MRM-MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Anal. Chem. 83, 2112–2118 (2011).
Prideaux, B. et al. Mass spectrometry imaging of levofloxacin distribution in TB-infected pulmonary lesions by MALDI-MSI and continuous liquid microjunction surface sampling. Int. J. Mass Spectrom. 377, 699–708 (2015).
Van Pelt, C. K. et al. A fully automated nanoelectrospray tandem mass spectrometric method for analysis of Caco-2 samples. Rapid Commun. Mass Spectrom. 17, 1573–1578 (2003).
Chen, C. Y., Lam, B. L. & Bhattacharya, S. K. Mass spectrometric analyses of phospholipids in the S334ter-3 rat model of retinal degeneration. Mol. Vis. 20, 1605–1611 (2014).
Yan, C. et al. Real-time screening of biocatalysts in live bacterial colonies. J. Am. Chem. Soc. 139, 1408–1411 (2017).
Wleklinski, M. et al. High throughput reaction screening using desorption electrospray ionization mass spectrometry. Chem. Sci. 9, 1647–1653 (2018).
Sinclair, I. et al. Novel acoustic loading of a mass spectrometer: toward next-generation high-throughput MS screening. J. Lab. Autom. 21, 19–26 (2016).
Heidari Torkabadi, H. et al. Following drug uptake and reactions inside Escherichia coli cells by Raman microspectroscopy. Biochemistry 53, 4113–4121 (2014).
Stratford, J. P. et al. Electrically induced bacterial membrane-potential dynamics correspond to cellular proliferation capacity. Proc. Natl Acad. Sci. USA 116, 9552–9557 (2019).
Cama, J., Henney, A. M. & Winterhalter, M. Breaching the barrier: quantifying antibiotic permeability across Gram-negative bacterial membranes. J. Mol. Biol. 431, 3531–3546 (2019).
Kuhn, P., Eyer, K., Allner, S., Lombardi, D. & Dittrich, P. S. A microfluidic vesicle screening platform: monitoring the lipid membrane permeability of tetracyclines. Anal. Chem. 83, 8877–8885 (2011).
Acknowledgements
We thank A. Duerfeldt and V. Rybenkov (University of Oklahoma); C. Balibar, D. McLaren, B. Sherborne, B. Squadroni, M. Tudor, and S. Walker (Merck Research Labs); H. Voss (Weill Cornell Medicine); and the entire SPEAR-GN team for helpful discussions. Financial support from the National Institutes of Health (R01 AI136795 to D.S.T. and H.I.Z.; R01 GM100477 and R01 AI118224 to D.S.T.; R01 AI136799 to H.I.Z.; and MSK CCSG P30 CA008748 to C. B. Thompson) is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
All authors contributed to the writing and editing of this manuscript. J.W.A. and H.I.Z. contributed Fig. 1. D.S.T. contributed Fig. 2. S.Z. and D.S.T. contributed Table 1. J.W.A., V.B., and H.I.Z. contributed Table 2.
Corresponding authors
Ethics declarations
Competing interests
Merck is a collaborating institution on the SPEAR-GN project and has provided in-kind support to the labs of D.S.T. and H.I.Z. D.S.T. serves on the External Advisory Board of the Institute for Research in Biomedicine, Barcelona; is a shareholder and has been a paid consultant and speaker for Merck; and has been a paid consultant or speaker for Eli Lilly, Elsevier, Emerson Collective, and Venenum Biosciences. H.I.Z. has been a paid speaker for Genentech and Novartis Research Institutes.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, S., Adamiak, J.W., Bonifay, V. et al. Defining new chemical space for drug penetration into Gram-negative bacteria. Nat Chem Biol 16, 1293–1302 (2020). https://doi.org/10.1038/s41589-020-00674-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-00674-6
This article is cited by
-
Multidrug efflux in Gram-negative bacteria: structural modifications in active compounds leading to efflux pump avoidance
npj Antimicrobials and Resistance (2024)
-
Unrealized targets in the discovery of antibiotics for Gram-negative bacterial infections
Nature Reviews Drug Discovery (2023)
-
Tackling the outer membrane: facilitating compound entry into Gram-negative bacterial pathogens
npj Antimicrobials and Resistance (2023)
-
Optimization, synthesis, and characterization of co-electrospinning cellulose acetate/thermoplastic polyurethanes composite membrane for photodynamic antibacterial application
Cellulose (2023)
-
A genetic platform to investigate the functions of bacterial drug efflux pumps
Nature Chemical Biology (2022)