Abstract
Lipids play crucial roles as structural elements, signaling molecules and material transporters in cells. However, the functions and dynamics of lipids within cells remain unclear because of a lack of methods to selectively label lipids in specific organelles and trace their movement by live-cell imaging. We describe here a technology for the selective labeling and fluorescence imaging (microscopic or nanoscopic) of phosphatidylcholine in target organelles. This approach involves the metabolic incorporation of azido-choline, followed by a spatially limited bioorthogonal reaction that enables the visualization and quantitative analysis of interorganelle lipid transport in live cells. More importantly, with live-cell imaging, we obtained direct evidence that the autophagosomal membrane originates from the endoplasmic reticulum. This method is simple and robust and is thus powerful for real-time tracing of interorganelle lipid trafficking.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and the Supplementary Information files. The materials and data reported in this study are available upon reasonable request from the corresponding author. Source data are provided with this paper.
References
Meer, G. Van, Voelker, D. R. & Feigenson, G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008).
Vance, J. E. Phospholipid synthesis and transport in mammalian cells. Traffic 16, 1–18 (2015).
Bumpus, T. W. & Baskin, J. M. Greasing the wheels of lipid biology with chemical tools. Trends Biochem. Sci. 43, 970–983 (2018).
Maekawa, M. & Fairn, G. D. Molecular probes to visualize the location, organization and dynamics of lipids. J. Cell Sci. 127, 4801–4812 (2014).
Yamaji, A. et al. Lysenin, a novel sphingomyelin-specific binding protein. J. Biol. Chem. 273, 5300–5306 (1998).
Kaplan, M. R. & Simoni, R. D. Intracellular transport of phosphatidylcholine to the plasma membrane. J. Cell Biol. 101, 441–445 (1985).
Takakura, H. et al. Long time-lapse nanoscopy with spontaneously blinking membrane probes. Nat. Biotechnol. 35, 773–780 (2017).
Gupta, A., Rivera-Molina, F., Xi, Z., Toomre, D. & Schepartz, A. Endosome motility defects revealed at super-resolution in live cells using HIDE probes. Nat. Chem. Biol. 16, 408–414 (2020).
Jao, C. Y., Roth, M., Welti, R. & Salic, A. Biosynthetic labeling and two-color imaging of phospholipids in cells. ChemBioChem 16, 472–476 (2015).
Neef, A. B. & Schultz, C. Selective fluorescence labeling of lipids in living cells. Angew. Chem. Int. Ed. 48, 1498–1500 (2009).
Bumpus, T. W. & Baskin, J. M. Clickable substrate mimics enable imaging of phospholipase D activity. ACS Cent. Sci. 3, 1070–1077 (2017).
Fujisawa, A., Tamura, T., Yasueda, Y., Kuwata, K. & Hamachi, I. Chemical profiling of the endoplasmic reticulum proteome using designer labeling reagents. J. Am. Chem. Soc. 140, 17060–17070 (2018).
Tamura, T. & Hamachi, I. Chemistry for covalent modification of endogenous/native proteins: from test tubes to complex biological systems. J. Am. Chem. Soc. 141, 2782–2799 (2019).
Alberts, B. et al. Molecular Biology of the Cell 5th edn (Garland Science, 2008).
Yasueda, Y. et al. A set of organelle-localizable reactive molecules for mitochondrial chemical proteomics in living cells and brain tissues. J. Am. Chem. Soc. 138, 7592–7602 (2016).
Kawano, S. et al. Structure–function insights into direct lipid transfer between membranes by Mmm1–Mdm12 of ERMES. J. Cell Biol. 217, 959–974 (2018).
Murase, K. et al. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys. J. 86, 4075–4093 (2004).
Emaus, R. K., Grunwald, R. & Lemasters, J. J. Rhodamine 123 as a probe of transmembrane potential in isolated rat-liver mitochondria: spectral and metabolic properties. Biochim. Biophys. Acta 850, 436–448 (1986).
Gao, W., Pu, Y., Luo, K. Q. & Chang, D. C. Temporal relationship between cytochrome c release and mitochondrial swelling during UV-induced apoptosis in living HeLa cells. J. Cell Sci. 114, 2855–2862 (2001).
Horvath, S. E. & Daum, G. Lipids of mitochondria. Prog. Lipid Res. 52, 590–614 (2013).
Haoxing, X. & Dejian, R. Lysosomal physiology. Annu. Rev. Physiol. 77, 57–80 (2015).
Matheoud, D. et al. Parkinson’s disease-related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell 166, 314–327 (2016).
Johannes, L., Tenza, D., Antony, C. & Goud, B. Retrograde transport of KDEL-bearing B-fragment of Shiga toxin. J. Biol. Chem. 272, 19554–19561 (1997).
Valm, A. M. et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 (2017).
Wood, S. A., Park, J. E. & Brown, W. J. Brefeldin A causes a microtubule-mediated fusion of the trans-Golgi network and early endosomes. Cell 67, 591–600 (1991).
Lamb, C. A., Yoshimori, T. & Tooze, S. A. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774 (2013).
Hailey, D. W. et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667 (2010).
Tooze, S. A. Current views on the source of the autophagosome membrane. Essays Biochem. 55, 29–38 (2013).
Chan, L. L. et al. A novel image-based cytometry method for autophagy detection in living cells. Autophagy 8, 1371–1382 (2012).
Munafó, D. B. & Colombo, M. I. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J. Cell Sci. 114, 3619–3629 (2001).
Tan, X., Thapa, N., Liao, Y., Choi, S. & Anderson, R. A. PtdIns(4,5)P2 signaling regulates ATG14 and autophagy. Proc. Natl Acad. Sci. USA 113, 10896–10901 (2016).
Hamasaki, M. et al. Autophagosomes form at ER–mitochondria contact sites. Nature 495, 389–393 (2013).
Hayashi-nishino, M. et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 (2009).
Axe, E. L. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 (2008).
Wakayama, S. et al. Chemical labelling for visualizing native AMPA receptors in live neurons. Nat. Commun. 8, 14850 (2017).
Kiyonaka, S. et al. Allosteric activation of membrane-bound glutamate receptors using coordination chemistry within living cells. Nat. Chem. 8, 958–967 (2016).
Miki, T. et al. A conditional proteomics approach to identify proteins involved in zinc homeostasis. Nat. Methods 13, 931–937 (2016).
Yoshii, T., Mizusawa, K., Takaoka, Y. & Hamachi, I. Intracellular protein-responsive supramolecules: protein sensing and in-cell construction of inhibitor assay system. J. Am. Chem. Soc. 136, 16635–16642 (2014).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Horger, K. S., Estes, D. J., Capone, R. & Mayer, M. Films of agarose enable rapid formation of giant liposomes in solutions of physiologic ionic strength. J. Am. Chem. Soc. 131, 1810–1819 (2009).
Ikenouchi, J. et al. Lipid polarity is maintained in absence of tight junctions. J. Biol. Chem. 287, 9525–9533 (2012).
Pantazaka, E. & Taylor, C. W. Differential distribution, clustering, and lateral diffusion of subtypes of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 286, 23378–23387 (2011).
Acknowledgements
We thank E. Kusaka (Kyoto University) for experimental support with NMR measurements. Super-resolution imaging was performed at the iCeMS Analysis Center, Institute for Integrated Cell-Material Sciences (iCeMS), Kyoto University Institute for Advanced Study (KUIAS). The electron microscopy study was supported by K. Okamoto-Furuta and H. Kohda (Division of Electron Microscopic Study, Center for Anatomical Studies, Graduate School of Medicine, Kyoto University). This work was funded by a Grant-in-Aid for Young Scientists (18K14334) and a Grant-in-Aid for Scientific Research on Innovative Areas ‘Integrated Bio-metal Science’ (19H05764) to T.T., the Japan Science and Technology Agency (JST) ERATO grant no. JPMJER1802 and by a Grant-in-Aid for Scientific Research on Innovative Areas ‘Chemistry for Multimolecular Crowding Biosystems’ (17H06348) to I.H. We thank J. Allen from the Edanz Group (https://www.edanzediting.com/ac) and M. Takato (Kyoto University) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
T.T., I.H. and A.F. conceived the idea and directed the project. T.T., A.F., M.T. and Y.S. conducted experiments. K.N. and M.U. supervised LC–MS/MS experiments and provided advice on data analysis. S.K., Y.T. and T.E. helped to conduct lipid transfer experiments. T.T. and I.H. prepared the manuscript with feedback from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
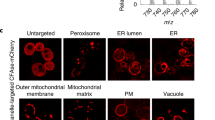
Extended Data Fig. 1 Product analysis of ER/Golgi-selective phosphatidylcholine (PC) labelling in live cells.
a, Thin-layer chromatography (TLC) analysis of the reaction products of 1 in N3-Cho-treated or nontreated cells. Eluent, chloroform/methanol/28% ammonia solution = 14/5/1. Lipid molecules were stained with cerium-ammonium-molybdate (CAM) solution. The fluorescence image was obtained before CAM stain. PC, phosphatidylcholine; PE, phosphatidylethanolamine; TAG, triacylglycerol. b, Mass spectrum of the labelled products indicated in (b). The labelled products on TLC were extracted and then subjected to liquid chromatography-mass spectrometry (LC-MS). There were no peaks derived from labelled sphingomyelins (SMs), ether PCs, or lyso PCs. All experiments shown are representative of at least two independent measurements.
Extended Data Fig. 2 ER/Golgi-specific PC labelling with 1.
a, Time-lapse imaging of washout process of unreacted 1 in HeLa cells. N3-Cho-treated or nontreated cells were incubated with 1 (100 nM) for 15 min. After washing with fresh medium, the cells were further incubated and analysed by CLSM at the indicated time points. Scale bars, 20 μm. b, SDS-PAGE analysis of the reactivity of 1 with proteins. N3-Cho-treated HeLa cells were incubated with 1 (100 nM) for 15 min, lysed, and analysed by SDS-PAGE. As a control experiment, ERM 6, an ER-localizable protein modification reagent previously developed by our group12, was used. c, Z-stack images of labelled HeLa cells. d–f, ER/Golgi-specific PC labelling in (d) A549 cells, (e) HEK293T cells, and (f) neurons. The labelling was performed using the same protocol as for HeLa cells. Scale bars, 5 μm. All experiments shown are representative of at least two independent measurements.
Extended Data Fig. 3 Labelling and imaging of PCs in mitochondria with 2.
a, TLC analysis of the reaction products of 2 in N3-Cho-treated or nontreated cells. Eluent, chloroform/methanol/28% ammonia solution = 14/5/1. b, Representative structure of 2-PC (16:0/18:1). c, MS spectrum of the labelled products of 2. The labelled product on TLC was extracted and then subjected to LC-MS. Similar to the ER-lipid labelling, there were no peaks derived from labelled SMs, ether PCs, or lyso PCs. d, Time-lapse imaging of washout process of unreacted 2 in HeLa cells. N3-Cho-treated or nontreated cells were incubated with 2 (100 nM) for 15 min. After washing with fresh medium, the cells were further incubated in culture medium and analysed by CLSM at the indicated time point. From this experiment, we determined the optimal washout time for 2 to be 1 h. Scale bars, 20 μm. e, SDS-PAGE analysis of the reactivity of 2 with proteins. N3-Cho-treated HeLa cells were incubated with 2 (100 nM) for 15 min, lysed, and analysed by SDS-PAGE. As a control experiment, MRM 1a, a mitochondria-localizable protein modification reagent previously developed by our group15, was used. f, g, Mitochondria-specific PC labelling in (f) A549 cells and (g) neurons (neurites). Scale bars, 5 μm. All experiments shown are representative of at least two independent measurements.
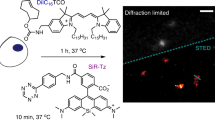
Extended Data Fig. 4 Super resolution imaging of submitochondrial localization of 2-PCs (magenta) with Zeiss LSM880 with Airyscan super-resolution imaging system in a fixed cell.
mEmerald-TOMM20 (green) was used to visualize the outer membrane of mitochondria. Scale bar, 1 µm. This data is representative of at least two independent measurements.
Extended Data Fig. 5 Immuno-electron microscopy of HeLa cells labelled with 2.
The labelled cells were fixed, sectioned on an ultramicrotome and stained with anti-tetraethylrhodamine antibody (primary Ab) and anti-rabbit IgG-10 nm gold conjugate (secondary Ab), counterstained with uranyl acetate and lead citrate, and imaged by transmission electron microscopy. a, Representative image of HeLa cells labeled with N3-Cho, followed by 2. Mt, mitochondria. b–d, Enlarged images of the area enclosed by the black square in (a). Yellow and red arrowheads indicate gold nano-particles observed in the mitochondrial outer and inner membrane, respectively. In (c), two gold nano-particles were observed around 30–35 nm (corresponding to the sum of the length of primary and secondary Abs) from the mitochondrial outer membrane (cytosolic leaflet), which indicates that the labelling can occur on the cytosolic leaflet of mitochondria. e, Representative images of negative control samples, in which only N3-Cho treatment was omitted in the lipid labelling process. No gold nanoparticles were detected. All experiments shown are representative of at least two independent measurements.
Extended Data Fig. 6 Translocation of 1-PCs from the ER/Golgi to other organelle membranes.
a, Evaluation of the intracellular stability of 1-PCs. After labelling, the cells were incubated in fresh medium for the indicated time period and subsequently analysed by TLC. This experiment confirmed that, the intracellular amount of 1-PCs did not alter in the experimental time range, and thus our observation is surely based on the labelled PCs itself. b, Time-lapse images of 1-PCs and ER-Tracker in HeLa cells. In this experiment, the signal intensities of 1-PCs were adjusted to be the same in all images. Scale bar, 5 μm. c, Quantification of the colocalization between 1-PCs and ER-Tracker in (b) (mean ± SD, n = 8). d–f, Translocation of 1-PCs from ER/Golgi to (d) lysosomes, (e) mitochondria, and (f) the PM. Yellow arrowheads indicate 1-PCs were transferred from the ER/Golgi to the other organelle by the indicated time. Scale bars, 5 μm, 0.5 µm and 2 µm for (d), (e, f), respectively. All experiments shown are representative of at least two independent measurements.
Extended Data Fig. 7 Translocation of 2-PCs from mitochondria to lysosomes.
a, Time-lapse images of 2-PCs and organelle markers in HeLa cells. Scale bar, 5 μm (b) Enlarged images of the region surrounded by white squares in (a). Lysosomal transport of 2-PCs was only observed 1320 min after labelling (yellow arrows). Scale bar, 2 μm. c, Evaluation of the intracellular stability of 2-PCs. After labelling, cells were incubated in fresh medium for the indicated time periods and subsequently analysed by TLC. d, Confocal images of 2-PCs and organelle markers in HeLa cells treated by heat-shock. After labelling with 2 at 37 °C, the cells were incubated at 37 °C (control) or 42 °C (heated) for 45 min. Yellow arrows indicate 2-PCs colocalized with lysosomes but not mitochondria. Scale bar, 5 μm. e, Heat-induced 2-PC translocation to lysosomes was evaluated by counting the number of lysosomal vesicles merged with 2-PCs in (d). p = 0.005 (two-tailed Student’s t-test). n = 3 and 5 cells for control and heated cells, respectively. Bar graphs show mean ± SD. All experiments shown are representative of at least two independent measurements.
Extended Data Fig. 8 Effects of Noc and BFA on interorganelle lipid transport.
a, Morphological abnormalities of the Golgi after Noc and BFA treatment. These data validated that microtubule disruption by Noc and collapse of the Golgi apparatus into the ER caused by BFA happen in our experimental conditions. These data shown are representative of at least two independent measurements. b, Time profiles of fluorescence decay of 1-PCs in the drug-treated cells. After labelling, the cells were treated with drugs for 30 min, followed by CLSM analysis. NS, not significant by two-tailed Student’s t-test. The graph shows mean ± SD. n = 33, 34, 41, 36, 34, 39, 37, 29, 42 cells for DMSO-30 min, −120 min, −240 min, Noc-30 min,−120 min, −240 min, BFA-30 min, −120 min, −240 min, respectively.
Extended Data Fig. 9 Interorganelle lipid transport is affected by treatment of nocodazole (Noc) and brefeldin A (BFA).
a, Confocal images of 1-PCs and organelle markers in HeLa cells treated with dimethylsulfoxide (DMSO, as control), nocodazole (Noc, 1 μM), or brefeldin A (BFA, 20 μM) for 4 h. Scale bar, 5 µm. These data shown are representative of at least two independent measurements. (b, c) Colocalization analysis of (a) with PCC value of (b) 1-PCs vs. ER/Golgi (ER-Tracker), and (c) 1-PCs vs. Lysosome (Lyso-Tracker). p-values shown were calculated by two-tailed Student’s t-test. Bar graphs represent mean ± S.E.M. The sample number is indicated in the graph.
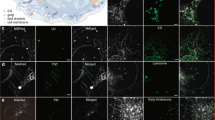
Extended Data Fig. 10 Translocation of 1-PCs from the ER to AP membranes.
a, Time-lapse images of starved HeLa cells. Colocalization of 1-PCs with LC3B-RFP is indicated by white vesicles. ER-Tracker and KDEL-GFP scarcely colocalized with AP vesicles. Scale bar, 5 μm. b, Colocalization of 1-PCs with mCherry-ATG5. White arrows indicate phagophores visualized by mCherry-ATG5, which were colocalized with 1-PCs on the ER membrane and moved together for several minutes. Scale bar, 1 μm. All experiments shown are representative of at least two independent measurements.
Supplementary information
Supplementary Information
Supplementary Note and Supplementary Figs. 1–8
Supplementary Video 1
Super-resolution imaging of submitochondrial localization of 2–PC with Zeiss Elyra 7 with lattice SIM in a live cell.
Supplementary Video 2
Super-resolution imaging of submitochondrial localization of mito–GFP with Zeiss Elyra 7 with lattice SIM in a live cell.
Supplementary Video 3
The overlay of Supplementary Videos 1 and 2.
Source data
Source Data Extended Data Fig. 2
Unprocessed gels of Extended Data Fig. 2b.
Source Data Extended Data Fig. 3
Unprocessed gels of Extended Data Fig. 3e.
Rights and permissions
About this article
Cite this article
Tamura, T., Fujisawa, A., Tsuchiya, M. et al. Organelle membrane-specific chemical labeling and dynamic imaging in living cells. Nat Chem Biol 16, 1361–1367 (2020). https://doi.org/10.1038/s41589-020-00651-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-00651-z
This article is cited by
-
Post-click labeling enables highly accurate single cell analyses of glucose uptake ex vivo and in vivo
Communications Biology (2024)
-
Bioorthogonal Chemistry in Cellular Organelles
Topics in Current Chemistry (2024)
-
Light-activated tetrazines enable precision live-cell bioorthogonal chemistry
Nature Chemistry (2022)
-
Live-cell fluorescence spectral imaging as a data science challenge
Biophysical Reviews (2022)