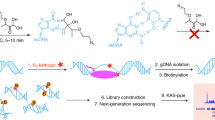
Abstract
RNA secondary structure is critical to RNA regulation and function. We report a new N3-kethoxal reagent that allows fast and reversible labeling of single-stranded guanine bases in live cells. This N3-kethoxal-based chemistry allows efficient RNA labeling under mild conditions and transcriptome-wide RNA secondary structure mapping.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All genomic data sets have been deposited in the Gene Expression Omnibus under accession number GSE122096. Other data and materials are available from the authors upon reasonable request.
Code availability
All custom codes used in this study are available at https://github.com/Tsinghua-gongjing/Keth-seq.
References
Wan, Y., Kertesz, M., Spitale, R. C., Segal, E. & Chang, H. Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 12, 641–655 (2011).
Kubota, M., Tran, C. & Spitale, R. C. Progress and challenges for chemical probing of RNA structure inside living cells. Nat. Chem. Biol. 11, 933–941 (2015).
Kertesz, M. et al. Genome-wide measurement of RNA secondary structure in yeast. Nature 467, 103–107 (2010).
Underwood, J. G. et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods 7, 995–1001 (2010).
Lucks, J. B. et al. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc. Natl Acad. Sci. USA 108, 11063–11068 (2011).
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M. & Weissman, J. S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705 (2014).
Ding, Y. et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700 (2014).
Talkish, J., May, G., Lin, Y., Woolford, J. L. & McManus, C. J. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA 20, 713–720 (2014).
Wan, Y. et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505, 706–709 (2014).
Spitale, R. C. et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490 (2015).
Zubradt, M. et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75–82 (2016).
Lu, Z. & Chang, H. Y. Decoding the RNA structurome. Curr. Opin. Struct. Biol. 36, 142–148 (2016).
National Toxicology Program. Dimethyl sulfate. Rep. Carcinog. 12, 174–175 (2011).
Merino, E. J., Wilkinson, K. A., Coughlan, J. L. & Weeks, K. M. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231 (2005).
Mitchell, D. et al. Glyoxals as in vivo RNA structural probes of guanine base-pairing. RNA 24, 114–124 (2018).
Mitchell, D. et al. In vivo RNA structural probing of uracil and guanine base pairing by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). RNA 25, 147–157 (2019).
Wang, P. Y., Sexton, A. N., Culligan, W. J. & Simon, M. D. Carbodiimide reagents for the chemical probing of RNA structure in cells. RNA 25, 135–146 (2019).
Feng, C. et al. Light-activated chemical probing of nucleobase solvent accessibility inside cells. Nat. Chem. Biol. 14, 276–283 (2018).
Xu, Z. & Culver, G.M. In Methods in Enzymology; Biophysical, Chemical, and Functional Probes of Rna Structure, Interactions and Folding, Pt A (ed. Herschalag, D.) Vol 468, 47–165 (Academic Press, 2009).
Morse, D. P. & Bass, B. L. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry 36, 8429–8434 (1997).
Andronescu, M., Bereg, V., Hoos, H. H. & Condon, A. RNA STRAND: the RNA secondary structure and statistical analysis database. BMC Bioinforma. 9, 340 (2008).
Kwok, C. K., Marsico, G., Sahakyan, A. B., Chambers, V. S. & Balasubramanian, S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods 13, 841–844 (2016).
Guo, J. U. & Bartel, D. P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 353, aaf5371 (2016).
Biffi, G., Di Antonio, M., Tannahill, D. & Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 6, 75–80 (2014).
Kwok, C. K., Marsico, G. & Balasubramanian, S. Detecting RNA G-quadruplexes (rG4s) in the transcriptome. Cold Spring Harb. Perspect. Biol. 10, a032284 (2018).
Spitale, R. C. et al. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 9, 18–20 (2013).
Lu, Z. et al. RNA Duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279 (2016).
Kalvari, I. et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46, D335–D442 (2018).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant nos. 21572172, 21778040 and 21822704 to X.W.; 21432008, 91753201 and 21721005 to X.Z.; 31671355, 91740204 and 31761163007 to Q.C.Z.) and National Institutes of Health grant no. HG008935 (C.H.). C.H. is an investigator of the Howard Hughes Medical Institute. X.W. was supported by China Scholarship Council (CSC) during his visit to the University of Chicago. We acknowledge S. Frank, who edited the manuscript.
Author information
Authors and Affiliations
Contributions
X.W., Q.C.Z., X.Z. and C.H. conceived the project, designed the experiments and wrote the manuscript. X.W., Y.C. and T.W. performed the experiments with the help of F.W., S.Y., Y.Y., G.L., K.C., L.H., H.M. and P.W. J.G. and Q.C.Z. designed and performed the bioinformatics analysis.
Corresponding authors
Ethics declarations
Competing interests
C.H. is a scientific founder and a member of the scientific advisory board of Accent Therapeutics, Inc., and a shareholder of Epican Genetech.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15 and Notes 1 and 2.
Rights and permissions
About this article
Cite this article
Weng, X., Gong, J., Chen, Y. et al. Keth-seq for transcriptome-wide RNA structure mapping. Nat Chem Biol 16, 489–492 (2020). https://doi.org/10.1038/s41589-019-0459-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0459-3
This article is cited by
-
KAS-seq profiling captures transcription dynamics during oocyte maturation
Journal of Ovarian Research (2024)
-
KARR-seq reveals cellular higher-order RNA structures and RNA–RNA interactions
Nature Biotechnology (2024)
-
Late-stage guanine C8–H alkylation of nucleosides, nucleotides, and oligonucleotides via photo-mediated Minisci reaction
Nature Communications (2024)
-
KARR-seq maps higher-order RNA structures and RNA–RNA interactions across the transcriptome
Nature Biotechnology (2024)
-
Integrated multiplexed assays of variant effect reveal determinants of catechol-O-methyltransferase gene expression
Molecular Systems Biology (2024)