Abstract
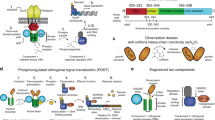
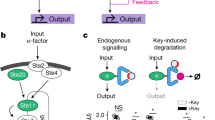
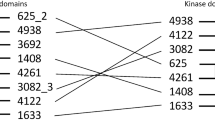
Augmenting live cells with new signal transduction capabilities is a key objective in genetic engineering and synthetic biology. We showed earlier that two-component signaling pathways could function in mammalian cells, albeit while losing their ligand sensitivity. Here, we show how to transduce small-molecule ligands in a dose-dependent fashion into gene expression in mammalian cells using two-component signaling machinery. First, we engineer mutually complementing truncated mutants of a histidine kinase unable to dimerize and phosphorylate the response regulator. Next, we fuse these mutants to protein domains capable of ligand-induced dimerization, which restores the phosphoryl transfer in a ligand-dependent manner. Cytoplasmic ligands are transduced by facilitating mutant dimerization in the cytoplasm, while extracellular ligands trigger dimerization at the inner side of a plasma membrane. These findings point to the potential of two-component regulatory systems as enabling tools for orthogonal signaling pathways in mammalian cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All plasmid sequences and data supporting the findings are available from the corresponding author upon request.
Code availability
The code used to process the time-lapse imaging data is available from the corresponding author upon request.
References
Park, S. H., Zarrinpar, A. & Lim, W. A. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299, 1061–1064 (2003).
Jackson, H. J., Rafiq, S. & Brentjens, R. J. Driving CAR T-cells forward. Nat. Rev. Clin. Oncol. 13, 370–383 (2016).
Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 13, 455–468 (2012).
Hansen, J. & Benenson, Y. Synthetic biology of cell signaling. Nat. Comput. 15, 5–13 (2016).
Cho, J. H., Collins, J. J. & Wong, W. W. Universal chimeric antigen receptors for multiplexed and logical control of T cell responses. Cell 173, 1426–1438 (2018).
Barnea, G. et al. The genetic design of signaling cascades to record receptor activation. Proc. Natl Acad. Sci. USA 105, 64–69 (2008).
Schwarz, K. A., Daringer, N. M., Dolberg, T. B. & Leonard, J. N. Rewiring human cellular input-output using modular extracellular sensors. Nat. Chem. Biol. 13, 202–209 (2017).
Kipniss, N. H. et al. Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system. Nat. Commun. 8, 2212 (2017).
Baeumler, T. A., Ahmed, A. A. & Fulga, T. A. Engineering synthetic signaling pathways with programmable dCas9-Based chimeric receptors. Cell Rep. 20, 2639–2653 (2017).
Xie, M. et al. Beta-cell-mimetic designer cells provide closed-loop glycemic control. Science 354, 1296–1301 (2016).
Porter, D. L., Levine, B. L., Kalos, M., Bagg, A. & June, C. H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733 (2011).
Mootz, H. D. & Muir, T. W. Protein splicing triggered by a small molecule. J. Am. Chem. Soc. 124, 9044–9045 (2002).
Slomovic, S. & Collins, J. J. DNA sense-and-respond protein modules for mammalian cells. Nat. Methods 12, 1085–1090 (2015).
Morsut, L. et al. Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell 164, 780–791 (2016).
Angelici, B., Mailand, E., Haefliger, B. & Benenson, Y. Synthetic biology platform for sensing and integrating endogenous transcriptional inputs in mammalian cells. Cell Rep. 16, 2525–2537 (2016).
Schreiber, J., Arter, M., Lapique, N., Haefliger, B. & Benenson, Y. Model‐guided combinatorial optimization of complex synthetic gene networks. Mol. Syst. Biol. 12, 899 (2016).
Hansen, J. et al. Transplantation of prokaryotic two-component signaling pathways into mammalian cells. Proc. Natl Acad. Sci. USA 111, 15705–15710 (2014).
Rivera-Cancel, G., Ko, W. H., Tomchick, D. R., Correa, F. & Gardner, K. H. Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation. Proc. Natl Acad. Sci. USA 111, 17839–17844 (2014).
Dago, A. E. et al. Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis. Proc. Natl Acad. Sci. USA 109, E1733–E1742 (2012).
Casino, P., Rubio, V. & Marina, A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139, 325–336 (2009).
T.N, H., Noriega, C. E. & Stewart, V. Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX. Proc. Natl Acad. Sci. Usa. 107, 21140–21145 (2010).
Landry, B. P., Palanki, R., Dyulgyarov, N., Hartsough, L. A. & Tabor, J. J. Phosphatase activity tunes two-component system sensor detection threshold. Nat. Commun. 9, 10 (2018).
Ashenberg, O., Rozen-Gagnon, K., Laub, M. T. & Keating, A. E. Determinants of homodimerization specificity in histidine kinases. J. Mol. Biol. 413, 222–235 (2011).
Tomomori, C. et al. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat. Struct. Biol. 6, 729–734 (1999).
Thompson, K. E., Bashor, C. J., Lim, W. A. & Keating, A. E. SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS Synth. Biol. 1, 118–129 (2012).
Cai, S. J. & Inouye, M. Spontaneous subunit exchange and biochemical evidence for trans-autophosphorylation in a dimer of Escherichia coli histidine kinase (EnvZ). J. Mol. Biol. 329, 495–503 (2003).
Casino, P., Miguel-Romero, L. & Marina, A. Visualizing autophosphorylation in histidine kinases. Nat. Commun. 5, 3258 (2014).
Cai, S. J. & Inouye, M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277, 24155–24161 (2002).
Bayle, J. H. et al. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem. Biol. 13, 99–107 (2006).
Schramm, A. et al. Establishing a split luciferase assay for proteinkinase G (PKG) interaction studies. Int. J. Mol. Sci. 19, 20 (2018).
Edwards, S. R. & Wandless, T. J. The rapamycin-binding domain of the protein kinase mammalian target of rapamycin is a destabilizing domain. J. Biol. Chem. 282, 13395–13401 (2007).
Kobilka, B. & Schertler, G. F. X. New G-protein-coupled receptor crystal structures: insights and limitations. Trends Pharmacol. Sci. 29, 79–83 (2008).
Billerbeck, S. et al. A scalable peptide-GPCR language for engineering multicellular communication. Nat. Commun. 9, 12 (2018).
Shukla, A. K. et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222 (2014).
Luttrell, L. M. & Lefkowitz, R. J. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell Sci. 115, 455–465 (2002).
Cahill, T. J. et al. Distinct conformations of GPCR-beta-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc. Natl Acad. Sci. USA 114, 2562–2567 (2017).
Baker, J. G. The selectivity of β‐adrenoceptor agonists at human β1‐, β2‐and β3‐adrenoceptors. Br. J. Pharmacol. 160, 1048–1061 (2010).
Kroeze, W. K. et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015).
O’Dowd, B. F., Hnatowich, M., Caron, M. G., Lefkowitz, R. J. & Bouvier, M. Palmitoylation of the human beta 2-adrenergic receptor. Mutation of Cys341 in the carboxyl tail leads to an uncoupled nonpalmitoylated form of the receptor. J. Biol. Chem. 264, 7564–7569 (1989).
Sadeghi, H. M., Innamorati, G., Dagarag, M. & Birnbaumer, M. Palmitoylation of the V2 vasopressin receptor. Mol. Pharmacol. 52, 21–29 (1997).
Klumpp, S. & Krieglstein, J. Reversible phosphorylation of histidine residues in proteins from vertebrates. Sci. Signal. 2, pe13 (2009).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Haefliger, B., Prochazka, L., Angelici, B. & Benenson, Y. Precision multidimensional assay for high-throughput microRNA drug discovery. Nat. Commun. 7, 12 (2016).
Pandy-Szekeres, G. et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 46, D440–D446 (2018).
Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988).
Acknowledgements
The study was funded by ETH Zurich, NCCR Molecular Systems Engineering (grant no. 51NF40-182895) and Swiss National Science foundation (grant no. 31003A_149802). We thank B. Haefliger, R. Altamura, J. Hansen, T. Littmann, G. Bernhardt and C. Stelzer for plasmids, Benenson lab members for discussions, H.M. Kaltenbach for help with statistical analysis and the members of the Single Cell Unit for their help with imaging and flow cytometry.
Author information
Authors and Affiliations
Contributions
A.M. conceived research, performed the experiments, analyzed data and wrote the paper. Y.B. conceived research, analyzed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
A patent application has been filed describing the result in this study, with A.M. and Y.B. listed as coinventors. Y.B. is a coinventor of a background patent to this filing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–27, Figs. 1–13 and Notes 1 and 2.
Rights and permissions
About this article
Cite this article
Mazé, A., Benenson, Y. Artificial signaling in mammalian cells enabled by prokaryotic two-component system. Nat Chem Biol 16, 179–187 (2020). https://doi.org/10.1038/s41589-019-0429-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0429-9
This article is cited by
-
Programmable synthetic receptors: the next-generation of cell and gene therapies
Signal Transduction and Targeted Therapy (2024)
-
Robust and tunable signal processing in mammalian cells via engineered covalent modification cycles
Nature Communications (2022)
-
Engineering the next generation of cell-based therapeutics
Nature Reviews Drug Discovery (2022)
-
Engineering living therapeutics with synthetic biology
Nature Reviews Drug Discovery (2021)
-
Molecular model of a sensor of two-component signaling system
Scientific Reports (2021)