Abstract
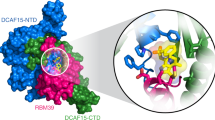
The anticancer agent indisulam inhibits cell proliferation by causing degradation of RBM39, an essential mRNA splicing factor. Indisulam promotes an interaction between RBM39 and the DCAF15 E3 ligase substrate receptor, leading to RBM39 ubiquitination and proteasome-mediated degradation. To delineate the precise mechanism by which indisulam mediates the DCAF15–RBM39 interaction, we solved the DCAF15–DDB1–DDA1–indisulam–RBM39(RRM2) complex structure to a resolution of 2.3 Å. DCAF15 has a distinct topology that embraces the RBM39(RRM2) domain largely via non-polar interactions, and indisulam binds between DCAF15 and RBM39(RRM2), coordinating additional interactions between the two proteins. Studies with RBM39 point mutants and indisulam analogs validated the structural model and defined the RBM39 α-helical degron motif. The degron is found only in RBM23 and RBM39, and only these proteins were detectably downregulated in indisulam-treated HCT116 cells. This work further explains how indisulam induces RBM39 degradation and defines the challenge of harnessing DCAF15 to degrade additional targets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the publication and its Supplementary Information or have been deposited in the PDB or Electron Microscopy Data Bank (http://www.ebi.ac.uk/pdbe/emdb/), as appropriate. Further information is available upon request. The PDB accession code for the human DCAF15–DDB1–DDA1–RBM39(RRM2)–indisulam EM co-structure is 6SJ7 and the EMDB accession code is EMD-10213. The PDB accession codes for the X-ray co-strutures of human DCAF15–DDB1(ΔBPB)–DDA1–RBM39(RRM2)–indisulam and human DCAF15–DDB1(ΔBPB)–DDA1–RBM39(RRM2)–compound 9 are 6UD7 and 6UE5, respectively.
Change history
15 January 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41589-020-0471-7
References
Bondeson, D. P. & Crew, C. M. Targeted protein degradation by small molecules. Annu. Rev. Pharmacol. Toxicol. 57, 107–123 (2017).
Collins, I., Wang, H., Caldwell, J. J. & Chopra, R. Chemical approaches to targeted protein degradation through modulation of the ubiquitin–proteasome pathway. Biochem. J. 474, 1127–1147 (2017).
Ciechanover, A. Intracellular protein degradation: from a vague Idea, through the lysosome and the ubiquitin–proteasome system, and onto human diseases and drug targeting (Nobel Lecture). Angew. Chem. Int. Ed. 44, 5944–5967 (2005).
Thrower, J. S. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 (2000).
Yu, H. & Matouschek, A. Recognition of client proteins by the proteasome. Annu. Rev. Biophys. 46, 149–173 (2017).
Finley, D., Chen, X. & Walters, K. J. Gates, channels, and switches: elements of the proteasome machine. Trends Biochem. Sci. 41, 77–93 (2016).
Neutzner, M. & Neutzner, A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 52, 37–50 (2012).
Zheng, N. & Shabek, N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017).
Clague, M. J., Heride, C. & Urbé, S. The demographics of the ubiquitin system. Trends Cell Biol. 25, 417–426 (2015).
Wu, Y. L. et al. Structural basis for an unexpected mode of SERM-Mediated ER antagonism. Mol. Cell 18, 413–424 (2005).
Patel, H. K. & Bihani, T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol. Therapeutics 186, 1–24 (2018).
Winkler, J. D., Neklesa, T. K. & Crews, C. M. Targeted protein degradation by PROTACs. Pharmacol. Ther. 174, 138–144 (2017).
Larrieu, A. & Vernoux, T. Comparison of plant hormone signalling systems. Essays Biochem. 58, 165–181 (2015).
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007).
Chamberlain, P. P. & Cathers, B. E. Cereblon modulators: low molecular weight inducers of protein degradation. Drug Discov. Today Technol. 31, 29–34 (2019).
Owa, T. et al. Discovery of novel antitumor sulfonamides targeting G1 phase of the cell cycle. J. Med. Chem. 42, 3789–3799 (1999).
Ozawa, Y. et al. E7070, a novel sulphonamide agent with potent antitumour activity in vitro and in vivo. Eur. J. Cancer 37, 2275–2282 (2001).
Uehara, T. et al. Selective degradation of splicing factor CAPERα by anticancer sulfonamides. Nat. Chem. Biol. 13, 675–680 (2017).
Han, T. et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, eaal3755 (2017).
Laskowski, R. A., Jabłońska, J., Pravda, L., Vařeková, R. S. & Thornton, J. M. PDBsum: structural summaries of PDB entries. Protein Sci. 27, 129–134 (2018).
Holm, L. & Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Dias, J. et al. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev. 28, 929–942 (2014).
Wysocka, J. et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859–872 (2005).
Song, J. J. & Kingston, R. E. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 283, 35258–35264 (2008).
Qu, Q. et al. Structure and conformational dynamics of a COMPASS histone H3K4 methyltransferase complex. Cell 174, 1117–1126 (2018).
Jain, B. P. & Pandey, S. WD40 repeat proteins: signalling scaffold with diverse functions. Protein J. 37, 391–406 (2018).
Xue, B., Dunbrack, R. L., Williams, R. W., Dunker, A. K. & Uversky, V. N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804, 996–1010 (2010).
Wu, Y. et al. The DDB1–DCAF1–Vpr–UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat. Struct. Mol. Biol. 23, 933–940 (2016).
Scrima, A. et al. Structural basis of UV DNA-damage recognition by the DDB1–DDB2 complex. Cell 135, 1213–1223 (2008).
Fischer, E. S. et al. Structure of the DDB1–CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014).
Shabek, N. et al. Structural insights into DDA1 function as a core component of the CRL4–DDB1 ubiquitin ligase. Cell Discov. 4, 67 (2018).
Chambers, J. C., Kenan, D., Martin, B. J. & Keene, J. D. Genomic structure and amino acid sequence domains of the human La autoantigen. J. Biol. Chem. 263, 18043–18051 (1988).
Dreyfuss, G., Swanson, M. S. & Piñol-Roma, S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem. Sci. 13, 86–91 (1988).
Murray, J. M. & Bussiere, D. E. Targeting the purinome. Methods Mol. Biol. 575, 47–92 (2009).
Molecular Operating Environment (MOE) 2018.01 (Chemical Computing Group ULC, Montreal, QC, Canada, 2018).
Milletti, F., Storchi, L., Sforna, G. & Cruciani, G. New and original pKa prediction method using grid molecular interaction fields. J. Chem. Inf. Model. 47, 2172–2181 (2007).
Kazlauskas, R. Engineering more stable proteins. Chem. Soc. Rev. 47, 9026–9045 (2018).
Pick, E. et al. Mammalian DET1 regulates CUL4A activity and forms stable complexes with E2 ubiquitin-conjugating enzymes. Mol. Cell. Biol. 27, 4708–4719 (2007).
Olma, M. H. et al. An interaction network of the mammalian COP9 signalosome identifies Dda1 as a core subunit of multiple Cul4-based E3 ligases. J. Cell Sci. 122, 1035–1044 (2009).
Sievers, Q. L. et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362, eaat0572 (2018).
Petzold, G., Fischer, E. S. & Thomä, N. H. Structural basis of lenalidomide-induced CK1α degradation by the CRL4 CRBN ubiquitin ligase. Nature 532, 127–130 (2016).
Matyskiela, M. E. et al. A novel cereblon modulator recruits GSPT1 to the CRL4 CRBN ubiquitin ligase. Nature 535, 252–257 (2016).
Laue, T. M., Shah, B. D., Ridgeway, T. M. & Pelletier, S. L. In Analytical Ultracentrifugation in Biochemistry and Polymer Science (Eds. Harding S. E. et al.) 90–125 (Royal Society of Chemistry, 1992).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Mayer, M. & Meyer, B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 123, 6108–6117 (2001).
Bhunia, A., Bhattacharjya, S. & Chatterjee, S. Applications of saturation transfer difference NMR in biological systems. Drug Discov. Today 17, 505–513 (2012).
Combe, C. W., Fischer, L. & Rappsilber, J. xiNET: cross-link network maps with residue resolution. Mol. Cell. Proteom. 14, 1137–1147 (2015).
Winter, G. Xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Winter, G. et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. Sect. D Struct. Biol. 74, 85–97 (2018).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 69, 1204–1214 (2013).
Adams, P. D.et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr.213–221 (2010).
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2210–2221 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 12–21 (2010).
Grant, T., Rohou, A. & Grigorieff, N. CisTEM, user-friendly software for single-particle image processing. Elife 7, e35383 (2018).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Allen, R. I., Box, K. J., Comer, J. E. A., Peake, C. & Tam, K. Y. Multiwavelength spectrophotometric determination of acid dissociation constants of ionizable drugs. J. Pharm. Biomed. Anal. 17, 699–712 (1998).
Erb, M. A. et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274 (2017).
Wessel, D. & Flügge, U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984).
Acknowledgements
The authors thank M. Renatus (Novartis) for providing DCAF15 constructs and M. Li (Novartis) for providing DDB1 and DDB1(ΔBPB) constructs. We also thank G. Pardee for baculovirus generation and protein expression, S. Widger for additional expression support, X. Ma for helpful discussions on ligase structural biology, S. Skolnik for supporting pKa measurements, and T. Rejtar for help with proteomics data informatics. We would like to thank T. Terwilliger and R. Read for the helpful discussions in regards to crystallographic molecular replacement and phasing. Finally, we thank J. Bradner, J. Shulok, R. Jain, J. Porter and J. Tallarico for helpful discussions, input on this manuscript and supporting this work.
Author information
Authors and Affiliations
Contributions
R.B., A. Fazal, J.Z., B.O., S.J. and P.-Y.M. designed and/or synthesized reported compounds. N.C., P.G. and H.V. performed crosslinking and mass spectrometry studies. A.B., D.K. and A.G performed SPR experiments. D.K. performed analytical ultracentrifugation. V.H. and R.G.K. performed proteome-wide motif searches and structural and computational modeling. C.B., H.S. and C.W. collected and processed cryo-EM data. F.X. and J.C. conducted expression proteomics experiments. A.O.F. and A. Frommlet performed biological NMR experiments. W.S. performed crystallographic screening and crystal optimization; D.E.B. designed protein constructs, collected X-ray crystallography datasets, reduced data, determined initial crystal structures and refined final structures; M.K. refined the final structures. L.X. purified DCAF15 complexes and RBM39 variants, and performed ITC and DSF experiments. J.P. and A.B. purified RBM39 variants, performed fluorescence-polarization and TR-FRET assays, cellular viability assays, siRNA knockdown and immunoblots. All authors contributed to writing. D.E.B., J.M.S., and J.P wrote and edited the final manuscript. D.E.B., J.M.S., L.X., and J.P contributed intellectual and strategic input.
Corresponding authors
Ethics declarations
Competing interests
All authors are employees of Novartis, or were at the time of this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information
Supplementary Tables 1 and 2 and Supplementary Figs. 1–16
Supplementary Note
Synthetic Procedures
Rights and permissions
About this article
Cite this article
Bussiere, D.E., Xie, L., Srinivas, H. et al. Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat Chem Biol 16, 15–23 (2020). https://doi.org/10.1038/s41589-019-0411-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0411-6
This article is cited by
-
DNA-encoded library-enabled discovery of proximity-inducing small molecules
Nature Chemical Biology (2024)
-
Targeted protein degradation via intramolecular bivalent glues
Nature (2024)
-
Applications and prospects of cryo-EM in drug discovery
Military Medical Research (2023)
-
RNA splicing dysregulation and the hallmarks of cancer
Nature Reviews Cancer (2023)
-
Molecular basis of RNA-binding and autoregulation by the cancer-associated splicing factor RBM39
Nature Communications (2023)