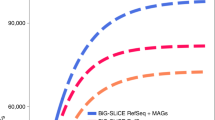
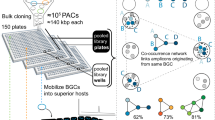
Abstract
Genome mining has become a key technology to exploit natural product diversity. Although initially performed on a single-genome basis, the process is now being scaled up to mine entire genera, strain collections and microbiomes. However, no bioinformatic framework is currently available for effectively analyzing datasets of this size and complexity. In the present study, a streamlined computational workflow is provided, consisting of two new software tools: the ‘biosynthetic gene similarity clustering and prospecting engine’ (BiG-SCAPE), which facilitates fast and interactive sequence similarity network analysis of biosynthetic gene clusters and gene cluster families; and the ‘core analysis of syntenic orthologues to prioritize natural product gene clusters’ (CORASON), which elucidates phylogenetic relationships within and across these families. BiG-SCAPE is validated by correlating its output to metabolomic data across 363 actinobacterial strains and the discovery potential of CORASON is demonstrated by comprehensively mapping biosynthetic diversity across a range of detoxin/rimosamide-related gene cluster families, culminating in the characterization of seven detoxin analogues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Genomes used in this study include assemblies from the sequencing project deposited in NCBI BioProject PRJNA488366, in Sequence Read Archive runs with accession numbers SRX4638772 to SRX4639021. AntiSMASH, BiG-SCAPE and CORASON results for all genome assemblies, along with raw files of phylogenetic trees, are available from ref. 50. Fully annotated nucleotide sequences for the BGCs for detoxin S1, detoxins N2–N3 and detoxins P1–P3 have been deposited in the Third Party Annotation Section of the DDBJ/ENA/GenBank databases under accession numbers BK010707, BK010852 and BK010851, respectively, and in MIBiG under accession numbers BGC0001840, BGC0001878 and BGC0001841, respectively. All raw MS data files for strains producing one or more of the nine compounds used for correlation analysis have been submitted to MassIVE under accession number MSV000083738. Raw MS data files and isolated MS/MS scan files for all newly identified detoxin analogues have been uploaded to MassIVE with accession number MSV000083648, and MS/MS data for other strains are available upon request.
Code availability
All our software is open source. An overview of both BiG-SCAPE and CORASON can be found at https://bigscape-corason.secondarymetabolites.org, BiG-SCAPE project at https://git.wur.nl/medema-group/BiG-SCAPE and CORASON project at https://github.com/nselem/corason.
References
Traxler, M. F. & Kolter, R. Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 32, 956–970 (2015).
Davies, J. Specialized microbial metabolites: functions and origins. J. Antibiot. 66, 361–364 (2013).
Cimermancic, P. et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 (2014).
Doroghazi, J. R. et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968 (2014).
Dejong, C. A. et al. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat. Chem. Biol. 12, 1007–1014 (2016).
Chevrette, M. G., Aicheler, F., Kohlbacher, O., Currie, C. R. & Medema, M. H. SANDPUMA: ensemble predictions of nonribosomal peptide chemistry reveal biosynthetic diversity across Actinobacteria. Bioinformatics 33, 3202–3210 (2017).
Pye, C. R., Bertin, M. J., Lokey, R. S., Gerwick, W. H. & Linington, R. G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl Acad. Sci. USA 114, 5601–5606 (2017).
Cruz-Morales, P. et al. Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model streptomycetes. Genome Biol. Evol. 8, 1906–1916 (2016).
Medema, M. H. & Fischbach, M. A. Computational approaches to natural product discovery. Nat. Chem. Biol. 11, 639–648 (2015).
Katz, L. & Baltz, R. H. Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol. 43, 155–176 (2016).
Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002).
Schneiker, S. et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 25, 1281–1289 (2007).
Bergmann, S. et al. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3, 213–217 (2007).
Medema, M. H. et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346 (2011).
Blin, K. et al. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41, W204–W212 (2013).
Weber, T. et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243 (2015).
Blin, K. et al. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41 (2017).
Johnston, C. W. et al. An automated Genomes-to-Natural Products platform (GNP) for the discovery of modular natural products. Nat. Commun. 6, 8421 (2015).
Skinnider, M. A. et al. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 43, 9645–9662 (2015).
Skinnider, M. A., Merwin, N. J., Johnston, C. W. & Magarvey, N. A. PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 45, W49–W54 (2017).
Medema, M. H. et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 11, 625–631 (2015).
Nielsen, J. C. et al. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat. Microbiol. 2, 17044 (2017).
Tobias, N. J. et al. Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat. Microbiol. 2, 1676–1685 (2017).
Grubbs, K. J. et al. Large-scale bioinformatics analysis of Bacillus genomes uncovers conserved roles of natural products in bacterial physiology. mSystems 2, e00040–17 (2017).
Freeman, M. F. et al. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 338, 387–390 (2012).
Agarwal, V. et al. Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat. Chem. Biol. 13, 537–543 (2017).
Owen, J. G. et al. Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. Proc. Natl Acad. Sci. USA 112, 4221–4226 (2015).
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017).
Leao, T. et al. Comparative genomics uncovers the prolific and distinctive metabolic potential of the cyanobacterial genus Moorea. Proc. Natl Acad. Sci. USA 114, 3198–3203 (2017).
Ziemert, N. et al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl Acad. Sci. USA 111, E1130–E1139 (2014).
Medema, M. H. et al. Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comput. Biol. 10, e1003822 (2014).
Mohimani, H. et al. NRPquest: coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J. Nat. Prod. 77, 1902–1909 (2014).
Mohimani, H. et al. Automated genome mining of ribosomal peptide natural products. ACS Chem. Biol. 9, 1545–1551 (2014).
Nguyen, D. D. et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl Acad. Sci. USA 110, E2611–E2620 (2013).
Goering, A. W. et al. Metabologenomics: correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent. Sci. 2, 99–108 (2016).
Duncan, K. R. et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem. Biol. 22, 460–471 (2015).
Punta, M. et al. The Pfam protein families databases. Nucleic Acids Res 40, D290–D301 (2012).
Medema, M. H., Cimermancic, P., Sali, A., Takano, E. & Fischbach, M. A. A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput. Biol. 10, e1004016 (2014).
Frey, B. J. & Dueck, D. Clustering by passing messages between data points. Science 315, 972–976 (2007).
Parkinson, E. I. et al. Discovery of the tyrobetaine natural products and their biosynthetic gene cluster via metabologenomics. ACS Chem. Biol. 13, 1029–1037 (2018).
McClure, R. A. et al. Elucidating the rimosamide-detoxin natural product families and their biosynthesis using metabolite/gene cluster correlations. ACS Chem. Biol. 11, 3452–3460 (2016).
Watrous, J. et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl Acad. Sci. USA 109, E1743–E1752 (2012).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009).
Hausinger, R. P. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem Mol. Biol. 39, 21–68 (2004).
Kim, K.-R., Kim, T.-J. & Suh, J.-W. The gene cluster for spectinomycin biosynthesis and the aminoglycoside-resistance function of spcM in Streptomyces spectabilis. Curr. Microbiol. 57, 371–374 (2008).
Sinha, A., Phillips-Salemka, S., Niraula, T.-A., Short, K. A. & Niraula, N. P. The complete genomic sequence of Streptomyces spectabilis NRRL-2792 and identification of secondary metabolite biosynthetic gene clusters. J. Ind. Microbiol Biotechnol. 46, 1217–1223 (2019).
Ogita, T., Seto, H., Otake, N. & Yonehara, H. The structures of minor congeners of the detoxin complex. Agric. Biol. Chem. 45, 2605–2611 (1981).
Yonehara, H., Seto, H., Aizawa, S., Hidaka, T. & Shimazu, A. The detoxin complex, selective antagonists of blasticidin S. J. Antibiot. (Tokyo) 21, 369–370 (1968).
Fischbach, M. A., Walsh, C. T. & Clardy, J. The evolution of gene collectives: how natural selection drives chemical innovation. Proc. Natl Acad. Sci. USA 105, 4601–4608 (2008).
Navarro-Muñoz J. C., Selem-Mojica N., Mullowney M. et al. Zenodo https://doi.org/10.5281/zenodo.1532752 (2018)..
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. InterJournal, Complex Systems 1695, 1–9 (2006).
Wickham, H. et al. ggplot2: an implementation of the grammar of graphics. R package version 7 http//CRANR-projectorg/package=ggplot2 (2008).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn Res. 12, 2825–2830 (2011).
Lin, K., Zhu, L. & Zhang, D. Y. An initial strategy for comparing proteins at the domain architecture level. Bioinformatics 22, 2081–2086 (2006).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Junier, T. & Zdobnov, E. M. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26, 1669–1670 (2010).
Henke, M. T. et al. New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans. ACS Chem. Biol. 11, 2117–2123 (2016).
Acknowledgements
We thank the following: the ARS of the USDA for providing bacterial strains; H. Sook Ann, Z. Crispino, Y. Kim, N. Ciszek and K. Espejo for generating bacterial culture extracts; R. McClure, M. Robey and G. Miley for assistance with and contributions to metabolomic data collection methods and acquisition; and Dr. Y. Zhang and Dr. Y. Wu of the Integrated Molecular Structure Education and Research Center (IMSERC) at Northwestern University for assistance in acquiring NMR data. Some analyses were carried out using CONABIO’s computing cluster, with funds from the Secretariat of Environment and Natural Resources. We thank K. Blin for technical assistance with setting up the website on the secondarymetabolites.org domain. The research reported in this publication was supported by the Netherlands Organization for Scientific Research (grant no. 863.15.002 to M.H.M.), the Graduate School for Experimental Plant Sciences (grant to M.H.M.); National Institutes of Health (NIH) Genome to Natural Products Network supplementary award (no. U01GM110706 to M.H.M.), CONACyT grants (grant nos. CBS2017_285746 and 2017_051TAMU to F.B.-G.; postdoctoral scholarship 263661 to J.C.N.M.; PhD scholarship 204482 to N.S.M. (who was also supported by the Innovation Secretary of Guanajuato)), the National Cancer Institute of the NIH (award no. F32CA221327 to M.W.M.), the National Institute of General Medical Sciences (award no. F32GM120999 to E.I.P.), the São Paulo Research Foundation (FAPESP, grant no. 17/08038-8 to L.T.D.C.), the National Center for Complementary and Integrative Health of the NIH (award no. R01AT009143 to R.J.T. and N.L.K.) and Warwick Integrative Synthetic Biology Centre, a UK Synthetic Biology Research grant from the Biotechnology and Biological Sciences Research Council and Engineering and Physical Sciences Research Council (grant no. BB/M017982/1 to E.L.C.D.L.S). This work made use of the IMSERC at Northwestern University, which has received support from the NIH (grant nos. 1S10OD012016-01/1S10RR019071-01A1), the State of Illinois and the International Institute for Nanotechnology. A.F.-G. received funding from the European Union’s Horizon 2020 research and innovation program (Blue Growth: Unlocking the Potential of Seas and Oceans; grant agreement no. 634486).
Author information
Authors and Affiliations
Contributions
R.J.T., W.W.M., N.L.K., F.B.-G. and M.H.M. originally conceived of the research and coordinated the work. J.C.N.M. designed and developed BiG-SCAPE, with the help of S.A.K., E.L.C.D.L.S., M.Y., S.A., A.R., W.L., A.F.-G. and M.H.M. S.A.K. designed the output visualizations with the help of J.C.N.M. and E.L.C.D.L.S. N.S.M. designed and developed CORASON, with the help of P.C.M. and F.B.-G. M.W.M., J.H.T., E.I.P., L.T.D.C., A.W.G., R.J.T., W.W.M. and N.L.K. designed and performed the experimental research. J.C.N.M., N.S.M., M.W.M., J.H.T., F.B.-G. and M.H.M. wrote the first draft of the manuscript and all authors participated in editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.H.M. is on the scientific advisory board of Hexagon Bio and co-founder of Design Pharmaceuticals. N.L.K., W.W.M. and R.J.T. are on the board of directors of MicroMGx, and A.W.G. is chief scientific officer at MicroMGx.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–7, Figs, 1–43 and Notes 1 and 2.
Supplementary Note 3
Synthetic Procedures
Rights and permissions
About this article
Cite this article
Navarro-Muñoz, J.C., Selem-Mojica, N., Mullowney, M.W. et al. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol 16, 60–68 (2020). https://doi.org/10.1038/s41589-019-0400-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0400-9
This article is cited by
-
Scoary2: rapid association of phenotypic multi-omics data with microbial pan-genomes
Genome Biology (2024)
-
Exploring shared therapeutic targets in diabetic cardiomyopathy and diabetic foot ulcers through bioinformatics analysis
Scientific Reports (2024)
-
Global characterization of biosynthetic gene clusters in non-model eukaryotes using domain architectures
Scientific Reports (2024)
-
bacLIFE: a user-friendly computational workflow for genome analysis and prediction of lifestyle-associated genes in bacteria
Nature Communications (2024)
-
A genome and gene catalog of the aquatic microbiomes of the Tibetan Plateau
Nature Communications (2024)