Abstract
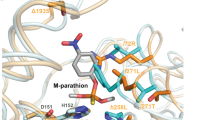
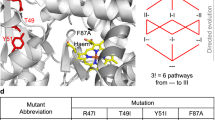
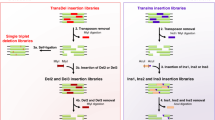
Characterizing the adaptive landscapes that encompass the emergence of novel enzyme functions can provide molecular insights into both enzymatic and evolutionary mechanisms. Here, we combine ancestral protein reconstruction with biochemical, structural and mutational analyses to characterize the functional evolution of methyl-parathion hydrolase (MPH), an organophosphate-degrading enzyme. We identify five mutations that are necessary and sufficient for the evolution of MPH from an ancestral dihydrocoumarin hydrolase. In-depth analyses of the adaptive landscapes encompassing this evolutionary transition revealed that the mutations form a complex interaction network, defined in part by higher-order epistasis, that constrained the adaptive pathways available. By also characterizing the adaptive landscapes in terms of their functional activities towards three additional organophosphate substrates, we reveal that subtle differences in the polarity of the substrate substituents drastically alter the network of epistatic interactions. Our work suggests that the mutations function collectively to enable substrate recognition via subtle structural repositioning.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The crystal structure of AncDHCH1 solved in this study has been deposited at the Protein Data Bank under accession code 6C2C. The raw data for the statistical analyses presented in Figs. 4, 5 and 6 and Supplementary Figs. 9 and 13 have been made publicly available at GitHub (https://github.com/danderson8/Yangetal2019.git). All other data supporting the findings of this study is available within the paper and its supplementary files.
Code availability
All analysis scripts, along with example data encoding, have been made publicly available via GitHub at https://github.com/danderson8/Yangetal2019.git.
Change history
12 June 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Weinreich, D. M., Delaney, N. F., Depristo, M. A. & Hartl, D. L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006).
Lozovsky, E. R. et al. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc. Natl Acad. Sci. USA 106, 12025–12030 (2009).
Sunden, F., Peck, A., Salzman, J., Ressl, S. & Herschlag, D. Extensive site-directed mutagenesis reveals interconnected functional units in the alkaline phosphatase active site. eLife 4, e06181 (2015).
Tufts, D. M. et al. Epistasis constrains mutational pathways of hemoglobin adaptation in high-altitude pikas. Mol. Biol. Evol. 32, 287–298 (2015).
Meini, M.-R., Tomatis, P. E., Weinreich, D. M. & Vila, A. J. Quantitative description of a protein fitness landscape based on molecular features. Mol. Biol. Evol. 32, 1774–1787 (2015).
Canale, A. S., Cote-Hammarlof, P. A., Flynn, J. M. & Bolon, D. N. Evolutionary mechanisms studied through protein fitness landscapes. Curr. Opin. Struct. Biol. 48, 141–148 (2018).
O’Maille, P. E. et al. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 4, 617–623 (2008).
Lunzer, M., Miller, S. P., Felsheim, R. & Dean, A. M. The biochemical architecture of an ancient adaptive landscape. Science 310, 499–501 (2005).
Clifton, B. E. et al. Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein. Nat. Chem. Biol. 14, 542–547 (2018).
Kaltenbach, M. et al. Evolution of chalcone isomerase from a non-catalytic ancestor. Nat. Chem. Biol. 14, 548–555 (2018).
Stormo, G. D. Maximally efficient modeling of DNA sequence motifs at all levels of complexity. Genetics 187, 1219–1224 (2011).
Anderson, D. W., McKeown, A. N. & Thornton, J. W. Intermolecular epistasis shaped the function and evolution of an ancient transcription factor and its DNA binding sites. eLife 4, e07864 (2015).
Weinreich, D. M., Lan, Y., Jaffe, J. & Heckendorn, R. B. The influence of higher-order epistasis on biological fitness landscape topography. J. Stat. Phys. 172, 208–225 (2018).
Sailer, Z. R. & Harms, M. J. High-order epistasis shapes evolutionary trajectories. PLoS Comput. Biol. 13, e1005541 (2017).
Sun, L. et al. Crystallization and preliminary X-ray studies of methyl parathion hydrolase from Pseudomonas sp. WBC-3. Acta Crystallogr. D 60, 954–956 (2004).
Malla, R. K., Bandyopadhyay, S., Spilling, C. D., Dutta, S. & Dupureur, C. M. The first total synthesis of (±)-cyclophostin and (±)-cyclipostin P: inhibitors of the serine hydrolases acetyl cholinesterase and hormone sensitive lipase. Org. Lett. 13, 3094–3097 (2011).
Nguyen, P. C. et al. Cyclipostins and cyclophostin analogs as promising compounds in the fight against tuberculosis. Sci. Rep. 7, 11751 (2017).
Liu, H., Zhang, J.-J., Wang, S.-J., Zhang, X.-E. & Zhou, N.-Y. Plasmid-borne catabolism of methyl parathion and p-nitrophenol in Pseudomonas sp. strain WBC-3. Biochem. Biophys. Res. Commun. 334, 1107–1114 (2005).
Luo, X. J. et al. Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp. Biotechnol. Bioeng. 111, 1920–1930 (2014).
Baier, F. & Tokuriki, N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 426, 2442–2456 (2014).
Khersonsky, O. & Tawfik, D. S. Structure–reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry 44, 6371–6382 (2005).
Purg, M. et al. Probing the mechanisms for the selectivity and promiscuity of methyl parathion hydrolase. Philos. Trans. A Math. Phys. Eng. Sci 374, 20160150 (2016).
Hong, S. B. & Raushel, F. M. Metal–substrate interactions facilitate the catalytic activity of the bacterial phosphotriesterase. Biochemistry 35, 10904–10912 (1996).
Jackson, C. J., Liu, J.-W., Coote, M. L. & Ollis, D. L. The effects of substrate orientation on the mechanism of a phosphotriesterase. Org. Biomol. Chem. 3, 4343–4350 (2005).
McKeown, A. N. et al. Evolution of DNA specificity in a transcription factor family produced a new gene regulatory module. Cell 159, 58–68 (2014).
Boucher, J. I., Jacobowitz, J. R., Beckett, B. C., Classen, S. & Theobald, D. L. An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases. eLife 3, e02304 (2014).
Bridgham, J. T., Carroll, S. M. & Thornton, J. W. Evolution of hormone-receptor complexity by molecular exploitation. Science 312, 97–101 (2006).
Kratzer, J. T. et al. Evolutionary history and metabolic insights of ancient mammalian uricases. Proc. Natl Acad. Sci. USA 111, 3763–3768 (2014).
Hochberg, G. K. A. & Thornton, J. W. Reconstructing ancient proteins to understand the causes of structure and function. Annu. Rev. Biophys. 46, 247–269 (2017).
Russell, R. J. et al. The evolution of new enzyme function: lessons from xenobiotic metabolizing bacteria versus insecticide-resistant insects. Evol. Appl. 4, 225–248 (2011).
Copley, S. D. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem. Sci. 25, 261–265 (2000).
Afriat-Jurnou, L., Jackson, C. J. & Tawfik, D. S. Reconstructing a missing link in the evolution of a recently diverged phosphotriesterase by active-site loop remodeling. Biochemistry 51, 6047–6055 (2012).
Crawford, R. L., Jung, C. M. & Strap, J. L. The recent evolution of pentachlorophenol (PCP)-4-monooxygenase (PcpB) and associated pathways for bacterial degradation of PCP. Biodegradation 18, 525–539 (2007).
Siddiq, M. A., Hochberg, G. K. & Thornton, J. W. Evolution of protein specificity: insights from ancestral protein reconstruction. Curr. Opin. Struct. Biol. 47, 113–122 (2017).
Starr, T. N., Picton, L. K. & Thornton, J. W. Alternative evolutionary histories in the sequence space of an ancient protein. Nature 549, 409–413 (2017).
Miton, C. M. & Tokuriki, N. How mutational epistasis impairs predictability in protein evolution and design. Protein Sci. 7, 1260–1272 (2016).
Kaltenbach, M. & Tokuriki, N. Dynamics and constraints of enzyme evolution. J. Exp. Zool. B Mol. Dev. Evol. 322, 468–487 (2014).
Koonin, E. V. Replaying the tape of life: quantification of the predictability of evolution. Front. Genet. 3, 246 (2012).
Ingles, D. W. & Knowles, J. R. Specificity and stereospecificity of alpha-chymotrypsin. Biochem. J. 104, 369–377 (1967).
Miton, C. M. et al. Evolutionary repurposing of a sulfatase: a new Michaelis complex leads to efficient transition state charge offset. Proc. Natl Acad. Sci. USA 115, E7293–E7302 (2018).
Jiménez-Osés, G. et al. The role of distant mutations and allosteric regulation on LovD active site dynamics. Nat. Chem. Biol. 10, 431–436 (2014).
Tokuriki, N. & Tawfik, D. S. Protein dynamism and evolvability. Science 324, 203–207 (2009).
Campbell, E. et al. The role of protein dynamics in the evolution of new enzyme function. Nat. Chem. Biol. 12, 944–950 (2016).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Wallace, I. M., O’Sullivan, O., Higgins, D. G. & Notredame, C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 34, 1692–1699 (2006).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008).
Abascal, F., Zardoya, R. & Posada, D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual 3rd edn (Spring Harbor Laboratory Press, 2001).
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 66, 133–144 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 64, 352–367 (2012).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Schrödinger, Release 2018-3 Maestro (Schrödinger, 2018).
Bas, D. C., Rogers, D. M. & Jensen, J. H. Very fast prediction and rationalization of pK a values for protein–ligand complexes. Proteins 73, 765–783 (2008).
Harder, E. et al. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 12, 281–296 (2016).
Friesner, R. A. et al. Extra precise glide: docking and scoring incorporating a model of hydrophobic enclosure for protein–ligand complexes. J. Med. Chem. 49, 6177–6196 (2006).
Jackson, C. J. et al. In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase. J. Mol. Biol. 375, 1189–1119 (2008).
Acknowledgements
We thank A. Pabis for performing computational analysis and providing revision and comments on the manuscript. N.T. and E.B.-B. thank the Human Frontier Science Program (HFSP) for support via research grant RGP0006/2013. N.T. acknowledges support by the Natural Sciences and Engineering Research Council of Canada (NSERC) via discovery grants RGPIN 418262-12 and RGPIN 2017-04909. N.T. is a CIHR new investigator and a Michael Smith Foundation of Health Research (MSFHR) career investigator. S.C.L.K. thanks the Knut and Alice Wallenberg Foundation (Wallenberg Academy Fellowships 2013.0124 and 2018.0140) and the Swedish National Infrastructure for Computing (SNIC). D.W.A. thanks NSERC and the MSFHR for post-doctoral support.
Author information
Authors and Affiliations
Contributions
G.Y. and N.T. conceived and designed this study. G.Y. and F.B. performed activity assays and mutational analysis. D.W.A. performed statistical analyses. E.B.-B. supervised bioinformatics. E.D. performed ancestral sequence reconstruction. F.B., N.H. and P.D.C. collected structural data under the supervision of C.J.J. N.H. and C.J.J. carried out molecular docking. S.C.L.K. designed computational analysis. G.Y. and N.T. wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–8 and Supplementary Figures 1–13.
Rights and permissions
About this article
Cite this article
Yang, G., Anderson, D.W., Baier, F. et al. Higher-order epistasis shapes the fitness landscape of a xenobiotic-degrading enzyme. Nat Chem Biol 15, 1120–1128 (2019). https://doi.org/10.1038/s41589-019-0386-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0386-3
This article is cited by
-
Opportunities and challenges in design and optimization of protein function
Nature Reviews Molecular Cell Biology (2024)
-
Epistasis arises from shifting the rate-limiting step during enzyme evolution of a β-lactamase
Nature Catalysis (2024)
-
Evolvability-enhancing mutations in the fitness landscapes of an RNA and a protein
Nature Communications (2023)
-
Pervasive epistasis exposes intramolecular networks in adaptive enzyme evolution
Nature Communications (2023)
-
Droplet-based screening of phosphate transfer catalysis reveals how epistasis shapes MAP kinase interactions with substrates
Nature Communications (2022)