Abstract
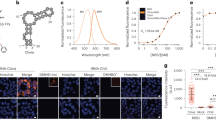
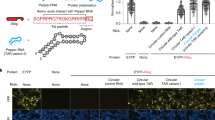
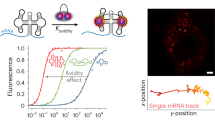
Live-cell imaging of RNA has remained a challenge because of the lack of naturally fluorescent RNAs. Recently developed RNA aptamers that can light-up small fluorogenic dyes could overcome this limitation, but they still suffer from poor brightness and photostability. Here, we propose the concept of a cell-permeable fluorogenic dimer of self-quenched sulforhodamine B dyes (Gemini-561) and the corresponding dimerized aptamer (o-Coral) that can drastically enhance performance of the current RNA imaging method. The improved brightness and photostability, together with high affinity of this complex, allowed direct fluorescence imaging in live mammalian cells of RNA polymerase III transcription products as well as messenger RNAs labeled with a single copy of the aptamer; that is, without tag multimerization. The developed fluorogenic module enables fast and sensitive detection of RNA inside live cells, while the proposed design concept opens the route to new generation of ultrabright RNA probes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information file.
References
Levine, J. H., Lin, Y. & Elowitz, M. B. Functional roles of pulsing in genetic circuits. Science 342, 1193–1200 (2013).
Giepmans, B. N., Adams, S. R., Ellisman, M. H. & Tsien, R. Y. The fluorescent toolbox for assessing protein location and function. Science 312, 217–224 (2006).
Xue, L., Karpenko, I. A., Hiblot, J. & Johnsson, K. Imaging and manipulating proteins in live cells through covalent labeling. Nat. Chem. Biol. 11, 917–923 (2015).
Dean, K. M. & Palmer, A. E. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat. Chem. Biol. 10, 512–523 (2014).
Bertrand, E. et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998).
Tutucci, E. et al. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods 15, 81–89 (2018).
Buxbaum, A. R., Haimovich, G. & Singer, R. H. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 16, 95–109 (2015).
Bouhedda, F., Autour, A. & Ryckelynck, M. Light-Up RNA aptamers and their cognate fluorogens: from their development to their applications. Int J. Mol. Sci. 19, 44 (2018).
Li, C., Tebo, A. G. & Gautier, A. Fluorogenic labeling strategies for biological imaging. Int. J. Mol. Sci. 18, 1473 (2017).
Klymchenko, A. S. Solvatochromic and fluorogenic dyes as environment-sensitive probes: design and biological applications. Acc. Chem. Res 50, 366–375 (2017).
Zhang, J. et al. Tandem spinach array for mrna imaging in living bacterial cells. Sci. Rep. 5, 17295 (2015).
Babendure, J. R., Adams, S. R. & Tsien, R. Y. Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc. 125, 14716–14717 (2003).
You, M. & Jaffrey, S. R. Structure and mechanism of RNA mimics of green fluorescent protein. Annu. Rev. Biophys. 44, 187–206 (2015).
Paige, J. S., Wu, K. Y. & Jaffrey, S. R. RNA mimics of green fluorescent protein. Science 333, 642–646 (2011).
Strack, R. L., Disney, M. D. & Jaffrey, S. R. A superfolding spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods 10, 1219–1224 (2013).
Filonov, G. S., Moon, J. D., Svensen, N. & Jaffrey, S. R. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 136, 16299–16308 (2014).
Song, W., Strack, R. L., Svensen, N. & Jaffrey, S. R. Plug-and-Play fluorophores extend the spectral properties of spinach. J. Am. Chem. Soc. 136, 1198–1201 (2014).
Han, K. Y., Leslie, B. J., Fei, J., Zhang, J. & Ha, T. Understanding the photophysics of the spinach-DFHBI RNA aptamer-fluorogen complex to improve live-cell RNA imaging. J. Am. Chem. Soc. 135, 19033–19038 (2013).
Constantin, T. P. et al. Synthesis of new fluorogenic cyanine dyes and incorporation into RNA fluoromodules. Org. Lett. 10, 1561–1564 (2008).
Murata, A., Sato, S., Kawazoe, Y. & Uesugi, M. Small-molecule fluorescent probes for specific RNA targets. Chem. Commun. 47, 4712–4714 (2011).
Dolgosheina, E. V. et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol. 9, 2412–2420 (2014).
Tan, X. et al. Fluoromodules consisting of a promiscuous RNA aptamer and red or blue fluorogenic cyanine dyes: selection, characterization, and bioimaging. J. Am. Chem. Soc. 139, 9001–9009 (2017).
Arora, A., Sunbul, M. & Jaschke, A. Dual-colour imaging of RNAs using quencher- and fluorophore-binding aptamers. Nucleic Acids Res. 43, e144 (2015).
Sunbul, M. & Jaschke, A. Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew. Chem. Int. Ed. Engl. 52, 13401–13404 (2013).
Sunbul, M. & Jaschke, A. SRB-2: a promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Res. 46, e110 (2018).
Sparano, B. A. & Koide, K. A strategy for the development of small-molecule-based sensors that strongly fluoresce when bound to a specific RNA. J. Am. Chem. Soc. 127, 14954–14955 (2005).
Shin, I. et al. Live-cell imaging of Pol II promoter activity to monitor gene expression with RNA IMAGEtag reporters. Nucleic Acids Res. 42, e90 (2014).
Holeman, L. A., Robinson, S. L., Szostak, J. W. & Wilson, C. Isolation and characterization of fluorophore-binding RNA aptamers. Fold. Des. 3, 423–431 (1998).
Karpenko, I. A. et al. Fluorogenic squaraine dimers with polarity-sensitive folding as bright far-red probes for background-free bioimaging. J. Am. Chem. Soc. 137, 405–412 (2015).
Okamoto, A. ECHO probes: a concept of fluorescence control for practical nucleic acid sensing. Chem. Soc. Rev. 40, 5815–5828 (2011).
Song, W. et al. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol. 13, 1187–1194 (2017).
Warner, K. D. et al. A homodimer interface without base pairs in an RNA mimic of red fluorescent protein. Nat. Chem. Biol. 13, 1195–1201 (2017).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Gotrik, M. et al. Direct selection of fluorescence-enhancing RNA aptamers. J. Am. Chem. Soc. 140, 3583–3591 (2018).
Autour, A., Westhof, E. & Ryckelynck, M. iSpinach: a fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res. 44, 2491–2500 (2016).
Autour, A. et al. Fluorogenic RNA mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 9, 656 (2018).
Ryckelynck, M. et al. Using droplet-based microfluidics to improve the catalytic properties of RNA under multiple-turnover conditions. RNA 21, 458–469 (2015).
Shulov, I. et al. Fluorinated counterion-enhanced emission of rhodamine aggregates: ultrabright nanoparticles for bioimaging and light-harvesting. Nanoscale 7, 18198–18210 (2015).
Collot, M. et al. Calcium rubies: a family of red-emitting functionalizable indicators suitable for two-photon Ca2+ imaging. J. Am. Chem. Soc. 134, 14923–14931 (2012).
Despras, G. et al. H-Rubies, a new family of red emitting fluorescent pH sensors for living cells. Chem. Sci. 6, 5928–5937 (2015).
Duval, M. et al. in RNA Structure and Folding: Biophysical Techniques and Prediction Methods (eds Klostermeier, D. & Hammann, C.) 29–50 (De Gruyter, 2013).
Trachman, R. J. 3rd, Truong, L. & Ferre-D’Amare, A. R. Structural principles of fluorescent RNA aptamers. Trends Pharm. Sci. 38, 928–939 (2017).
Filonov, G. S., Kam, C. W., Song, W. & Jaffrey, S. R. In-gel imaging of RNA processing using Broccoli reveals optimal aptamer expression strategies. Chem. Biol. 22, 649–660 (2015).
Good, P. D. et al. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 4, 45–54 (1997).
Guet, D. et al. Combining spinach-tagged RNA and gene localization to image gene expression in live yeast. Nat. Commun. 6, 8882 (2015).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
Baugh, C., Grate, D. & Wilson, C. 2.8 A crystal structure of the malachite green aptamer. J. Mol. Biol. 301, 117–128 (2000).
Shelke, S. A. et al. Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif. Nat. Commun. 9, 4542 (2018).
Guo, J. U. & Bartel, D. P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 353, aaf5371 (2016).
Acknowledgements
We thank R. Leblay, A.-C. Helfer, E. Gutzwiller and G. Lieber for technical assistance as well as P. Romby and I. Caldelari for fruitful scientific discussion. We thank O. Kucherak for synthesis of intermediate 2, E. Real for scientific discussions and J. Valanciunaite and N. Humbert for the technical assistance. This work received financial support from Agence Nationale de la Recherche (BrightRiboProbes, grant no. ANR-16-CE11-0010-01/02 to M.R. and A.S.K.) and BacNet (grant no. ANR-10-BINF_02_02 to M.R.) and European Research Council ERC Consolidator grant BrightSens (no. 648528 to A.S.K.). This work has also been published under the framework of the LabEx (no. ANR-10-LABX-0036_NETRNA to M.R.) and benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the Future Program. It was also supported by the Université de Strasbourg and the Centre National de la Recherche Scientifique.
Author information
Authors and Affiliations
Contributions
A.K., M.C. and M.R. proposed the concept of this study. M.C. synthesized Gemini-561. F.B. and K.T.F. performed main part of experimental work and data analysis with help and supervision of M.R., M.C. and A.K. F.B. and M.R. developed the aptamer o-Coral, including its variants and characterized their structure with the help of S.M. A.A. and F.B. characterized the contribution of the different selected mutations. K.T.F., M.C., F.B. and A.K. characterized Gemini-561–o-Coral fluorescence properties in solution. F.B. generated plasmids encoding o-Coral. K.T.F. realized all cellular studies and all fluorescence imaging of the cells, with help of A.K. and M.C.
Corresponding authors
Ethics declarations
Competing interests
F.B., K.T.F., M.C., A.K., M.R., the University of Strasbourg and the CNRS have filed a patent application covering the technology presented in this manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information
Supplementary Tables 1–3 and Supplementary Figures 1–27.
Supplementary Note
Synthetic procedures
Rights and permissions
About this article
Cite this article
Bouhedda, F., Fam, K.T., Collot, M. et al. A dimerization-based fluorogenic dye-aptamer module for RNA imaging in live cells. Nat Chem Biol 16, 69–76 (2020). https://doi.org/10.1038/s41589-019-0381-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0381-8
This article is cited by
-
Fast-exchanging spirocyclic rhodamine probes for aptamer-based super-resolution RNA imaging
Nature Communications (2023)
-
Large Stokes shift fluorescent RNAs for dual-emission fluorescence and bioluminescence imaging in live cells
Nature Methods (2023)
-
Droplet-based microfluidics
Nature Reviews Methods Primers (2023)
-
Design and construction of a microfluidics workstation for high-throughput multi-wavelength fluorescence and transmittance activated droplet analysis and sorting
Nature Protocols (2023)
-
Avidity-based bright and photostable light-up aptamers for single-molecule mRNA imaging
Nature Chemical Biology (2023)