Abstract
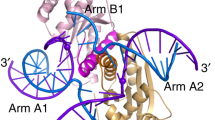
The Holliday junction (HJ) is a key intermediate during homologous recombination and DNA double-strand break repair. Timely HJ resolution by resolvases is critical for maintaining genome stability. The mechanisms underlying sequence-specific substrate recognition and cleavage by resolvases remain elusive. The monokaryotic chloroplast 1 protein (MOC1) specifically cleaves four-way DNA junctions in a sequence-specific manner. Here, we report the crystal structures of MOC1 from Zea mays, alone or bound to HJ DNA. MOC1 uses a unique β-hairpin to embrace the DNA junction. A base-recognition motif specifically interacts with the junction center, inducing base flipping and pseudobase-pair formation at the strand-exchanging points. Structures of MOC1 bound to HJ and different metal ions support a two-metal ion catalysis mechanism. Further molecular dynamics simulations and biochemical analyses reveal a communication between specific substrate recognition and metal ion-dependent catalysis. Our study thus provides a mechanism for how a resolvase turns substrate specificity into catalytic efficiency.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Atomic coordinates and structure factors of apo-MOC1, MOC1–HJ–Ca2+, MOC1D115N–HJ–Mg2+ and MOC1H253A–HJ–Mg2+ have been deposited in the Protein Data Bank (PDB) with accession codes 6IS9, 6JRF, 6IS8 and 6JRG, respectively. All data generated and analyzed during this study are included in this published article. Constructs encoding full-length and truncated ZmMOC1 variants are available from the corresponding author upon reasonable request.
References
Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. 5, 282–304 (1964).
Lilley, D. M. & White, M. F. The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2, 433–443 (2001).
Mizuuchi, K., Kemper, B., Hays, J. & Weisberg, R. A. T4 endonuclease VII cleaves holliday structures. Cell 29, 357–365 (1982).
Connolly, B. et al. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc. Natl Acad. Sci. USA 88, 6063–6067 (1991).
Iwasaki, H., Takahagi, M., Shiba, T., Nakata, A. & Shinagawa, H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 10, 4381–4389 (1991).
Dunderdale, H. J. et al. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature 354, 506–510 (1991).
Komori, K., Sakae, S., Shinagawa, H., Morikawa, K. & Ishino, Y. A holliday junction resolvase from Pyrococcus furiosus: functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in Bacteria, Eukarya, and Archaea. Proc. Natl Acad. Sci. USA 96, 8873–8878 (1999).
Kleff, S., Kemper, B. & Sternglanz, R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 11, 699–704 (1992).
Symington, L. S. & Kolodner, R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc. Natl Acad. Sci. USA 82, 7247–7251 (1985).
West, S. C., Parsons, C. A. & Picksley, S. M. Purification and properties of a nuclease from Saccharomyces cerevisiae that cleaves DNA at cruciform junctions. J. Biol. Chem. 262, 12752–12758 (1987).
White, M. F. & Lilley, D. M. Characterization of a holliday junction-resolving enzyme from Schizosaccharomyces pombe. Mol. Cell Biol. 17, 6465–6471 (1997).
Constantinou, A., Davies, A. A. & West, S. C. Branch migration and holliday junction resolution catalyzed by activities from mammalian cells. Cell 104, 259–268 (2001).
Boddy, M. N. et al. Mus81-Eme1 are essential components of a holliday junction resolvase. Cell 107, 537–548 (2001).
Ip, S. C. et al. Identification of holliday junction resolvases from humans and yeast. Nature 456, 357–361 (2008).
Wyatt, H. D. & West, S. C. Holliday junction resolvases. Cold Spring Harb. Perspect. Biol. 6, a023192 (2014).
Shah, R., Bennett, R. J. & West, S. C. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell 79, 853–864 (1994).
White, M. F. & Lilley, D. M. The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J. Mol. Biol. 257, 330–341 (1996).
Kobayashi, Y. et al. Holliday junction resolvases mediate chloroplast nucleoid segregation. Science 356, 631–634 (2017).
Chen, L., Shi, K., Yin, Z. & Aihara, H. Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during holliday junction resolution. Nucleic Acids Res. 41, 648–656 (2013).
Gorecka, K. M., Komorowska, W. & Nowotny, M. Crystal structure of RuvC resolvase in complex with holliday junction substrate. Nucleic Acids Res. 41, 9945–9955 (2013).
Biertumpfel, C., Yang, W. & Suck, D. Crystal structure of T4 endonuclease VII resolving a holliday junction. Nature 449, 616–620 (2007).
Hadden, J. M., Declais, A. C., Carr, S. B., Lilley, D. M. & Phillips, S. E. The structural basis of holliday junction resolution by T7 endonuclease I. Nature 449, 621–624 (2007).
Nowotny, M., Gaidamakov, S. A., Crouch, R. J. & Yang, W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016 (2005).
Steitz, T. A. & Steitz, J. A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 (1993).
Nowotny, M. & Yang, W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 25, 1924–1933 (2006).
Majorek, K. A. et al. The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 42, 4160–4179 (2014).
Yang, W., Lee, J. Y. & Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006).
Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993).
Wang, Q. -S et al. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 29, 68 (2018).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Case, D. A. et al. AMBER 2016 (University of California: San Francisco, CA, USA, 2016).
Gordon, J. C. et al. H++: a server for estimating pK(a)s and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33, W368–W371 (2005).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Ivani, I. et al. Parmbsc1: a refined force field for DNA simulations. Nat. Methods 13, 55–58 (2016).
Aqvist, J. Ion water interaction potentials derived from free-energy perturbation simulations. J. Phys. Chem. 94, 8021–8024 (1990).
Li, P. F. & Merz, K. M. Taking into account the ion-induced dipole interaction in the nonbonded model of ions. J. Chem. Theory Comput. 10, 289–297 (2014).
Panteva, M. T., Giambasu, G. M. & York, D. M. Comparison of structural, thermodynamic, kinetic and mass transport properties of Mg2+ ion models commonly used in biomolecular simulations. J. Comput. Chem. 36, 970–982 (2015).
Panteva, M. T., Giambasu, G. M. & York, D. M. Force field for Mg2+, Mn2+, Zn2+, and Cd2+ ions that have balanced interactions with nucleic acids. J. Phys. Chem. B 119, 15460–15470 (2015).
Sponer, J. et al. RNA structural dynamics as captured by molecular simulations: a comprehensive overview. Chem. Rev. 118, 4177–4338 (2018).
Li, P. F. & Merz, K. M. Metal ion modeling using classical mechanics. Chem. Rev. 117, 1564–1686 (2017).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Toukmaji, A., Sagui, C., Board, J. & Darden, T. Efficient particle-mesh ewald based approach to fixed and induced dipolar interactions. J. Chem. Phys. 113, 10913–10927 (2000).
Hoover, W. G. Canonical dynamics - equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 (1985).
Zhang, Y. H., Feller, S. E., Brooks, B. R. & Pastor, R. W. Computer-simulation of liquid/liquid interfaces. 1. Theory and application to octane/water. J. Chem. Phys. 103, 10252–10266 (1995).
Acknowledgements
We thank the staff of BL17B, BL18U1 and BL19U1 beamlines at the National Facility for Protein Science Shanghai (NFPS) and Shanghai Synchrotron Radiation Facility, Shanghai, People’s Republic of China, for assistance with X-ray data collection. We thank H. Yu (Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas, TX) for critical comments on this manuscript and Y. Huang (Fuzhou University, Fujian, China) for assistance with the molecular dynamics simulations. This work is supported by the National Natural Science Foundation of China 31971222 (Z.L.), 21603033 (J.L.), 31370737 (M.H.) and 31670739 (M.H.), and the National Key R&D Program of China 2017YFE0103200 (M.H.).
Author information
Authors and Affiliations
Contributions
H.L. carried out all the cloning, DNA binding and cleavage experiments, and the protein purification and crystallization of both native and SeMet MOC1. D.Z. carried out the purification and crystallization of MOC1–HJ complexes. H.L. and D.Z. performed the X-ray data collection. C.Y. performed structure refinements. K.Z. carried out the molecular dynamics simulations under the supervision of J.L. M.H. supervised the project and analyzed the data. Z.L. conceived and designed the project, determined the crystal structures and wrote the manuscript with the input of all authors. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–4, Supplementary Figs. 1–17
Rights and permissions
About this article
Cite this article
Lin, H., Zhang, D., Zuo, K. et al. Structural basis of sequence-specific Holliday junction cleavage by MOC1. Nat Chem Biol 15, 1241–1248 (2019). https://doi.org/10.1038/s41589-019-0377-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0377-4
This article is cited by
-
Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1
Nature Communications (2020)