Abstract
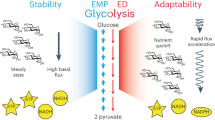
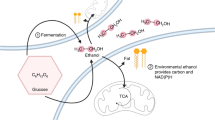
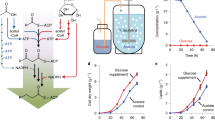
Glycolysis plays a central role in producing ATP and biomass. Its control principles, however, remain incompletely understood. Here, we develop a method that combines 2H and 13C tracers to determine glycolytic thermodynamics. Using this method, we show that, in conditions and organisms with relatively slow fluxes, multiple steps in glycolysis are near to equilibrium, reflecting spare enzyme capacity. In Escherichia coli, nitrogen or phosphorus upshift rapidly increases the thermodynamic driving force, deploying the spare enzyme capacity to increase flux. Similarly, respiration inhibition in mammalian cells rapidly increases both glycolytic flux and the thermodynamic driving force. The thermodynamic shift allows flux to increase with only small metabolite concentration changes. Finally, we find that the cellulose-degrading anaerobe Clostridium cellulolyticum exhibits slow, near-equilibrium glycolysis due to the use of pyrophosphate rather than ATP for fructose-bisphosphate production, resulting in enhanced per-glucose ATP yield. Thus, near-equilibrium steps of glycolysis promote both rapid flux adaptation and energy efficiency.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data for Figs. 1–6 are provided in Supplementary Tables 1–13 and on the GitHub public repository: https://github.com/jopark/GibbsIT
Code availability
The code for metabolic flux and free energy analysis is available on the GitHub public repository: https://github.com/jopark/GibbsIT
References
Tanner, L. B. et al. Four key steps control glycolytic flux in mammalian cells. Cell Syst. 7, 49–62.e48 (2018).
Henry, C. S., Broadbelt, L. J. & Hatzimanikatis, V. Thermodynamics-based metabolic flux analysis. Biophys. J. 92, 1792–1805 (2007).
Fell, D. Understanding the Control of Metabolism (Portland Press, 1997).
Hackett, S. R. et al. Systems-level analysis of mechanisms regulating yeast metabolic flux. Science 354, aaf2786 (2016).
Flamholz, A., Noor, E., Bar-Even, A., Liebermeister, W. & Milo, R. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc. Natl Acad. Sci. USA 110, 10039–10044 (2013).
Dona, A. C. et al. A guide to the identification of metabolites in NMR-based metabonomics/metabolomics experiments. Computat. Struct. Biotechnol. J. 14, 135–153 (2016).
Bennett, B. D., Yuan, J., Kimball, E. H. & Rabinowitz, J. D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 3, 1299–1311 (2008).
Lu, W. et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal. Chem. 82, 3212–3221 (2010).
Katz, L. A., Swain, J. A., Portman, M. A. & Balaban, R. S. Intracellular pH and inorganic phosphate content of heart in vivo: a 31P-NMR study. Am. J. Physiol. 255, H189–H196 (1988).
Lu, W. et al. Metabolite measurement: pitfalls to avoid and practices to follow. Annu. Rev. Biochem. 86, 277–304 (2017).
Noor, E., Haraldsdóttir, H. S., Milo, R. & Fleming, R. M. T. Consistent estimation of Gibbs energy using component contributions. PLoS Comput. Biol. 9, e1003098 (2013).
Du, B. et al. Temperature-dependent estimation of gibbs energies using an updated group-contribution method. Biophys. J. 114, 2691–2702 (2018).
Park, J. O. et al. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat. Chem. Biol. 12, 482–489 (2016).
Beard, D. A. & Qian, H. Relationship between thermodynamic driving force and one-way fluxes in reversible processes. PLoS One 2, e144 (2007).
Harris, T. K., Abeygunawardana, C. & Mildvan, A. S. NMR studies of the role of hydrogen bonding in the mechanism of triosephosphate isomerase. Biochemistry 36, 14661–14675 (1997).
Poyner, R. R., Laughlin, L. T., Sowa, G. A. & Reed, G. H. Toward identification of acid/base catalysts in the active site of enolase: comparison of the properties of K345A, E168Q, and E211Q variants. Biochemistry 35, 1692–1699 (1996).
Xu, Y.-F., Lu, W. & Rabinowitz, J. D. Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography–mass spectrometry-based metabolomics. Anal. Chem. 87, 2273–2281 (2015).
Antoniewicz, M. R., Kelleher, J. K. & Stephanopoulos, G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab. Eng. 9, 68–86 (2007).
Bren, A. et al. Glucose becomes one of the worst carbon sources for E. coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci. Rep. 6, 24834 (2016).
Doucette, C. D., Schwab, D. J., Wingreen, N. S. & Rabinowitz, J. D. α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 7, 894–901 (2011).
Yuan, J. et al. Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli. Mol. Syst. Biol. 5, 302–302 (2009).
Kustu, S., Hirschman, J., Burton, D., Jelesko, J. & Meeks, J. C. Covalent modification of bacterial glutamine-synthetase—physiological significance. Mol. Gen. Genet. 197, 309–317 (1984).
Ikeda, T. P., Shauger, A. E. & Kustu, S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259, 589–607 (1996).
Xu, Y.-F., Amador-Noguez, D., Reaves, M. L., Feng, X.-J. & Rabinowitz, J. D. Ultrasensitive regulation of anapleurosis via allosteric activation of PEP carboxylase. Nat. Chem. Biol. 8, 562–568 (2012).
Pike Winer, L. S. & Wu, M. Rapid analysis of glycolytic and oxidative substrate flux of cancer cells in a microplate. PLoS One 9, e109916 (2014).
Desvaux, M. Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol. Rev. 29, 741–764 (2005).
Zhou, J. L. et al. Atypical glycolysis in Clostridium thermocellum. Appl. Environ. Microbiol. 79, 3000–3008 (2013).
Chen, J. et al. Pyrophosphatase is essential for growth of Escherichia coli. J. Bacteriol. 172, 5686–5689 (1990).
Mertens, E. Pyrophosphate-dependent phosphofructokinase, an anaerobic glycolytic enzyme? FEBS Lett. 285, 1–5 (1991).
Beg, Q. K. et al. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc. Natl Acad. Sci. USA 104, 12663–12668 (2007).
Basan, M. et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104 (2015).
Scott, M., Gunderson, C. W., Mateescu, E. M., Zhang, Z. G. & Hwa, T. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 (2010).
Schuetz, R., Zamboni, N., Zampieri, M., Heinemann, M. & Sauer, U. Multidimensional optimality of microbial metabolism. Science 336, 601–604 (2012).
Dekel, E. & Alon, U. Optimality and evolutionary tuning of the expression level of a protein. Nature 436, 588–592 (2005).
Tian, L. et al. Metabolome analysis reveals a role for glyceraldehyde 3-phosphate dehydrogenase in the inhibition of C. thermocellum by ethanol. Biotechnol. Biofuels 10, 276 (2017).
Heinrich, R. & Schuster, S. The Regulation of Cellular Systems (Chapman & Hall, 1996).
Hofmeyr, J. H. & Cornish-Bowden, A. Quantitative assessment of regulation in metabolic systems. Eur. J. Biochem. 200, 223–236 (1991).
Gutnick, D., Calvo, J. M., Klopotow., T. & Ames, B. N. Compounds which serve as sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 100, 215–219 (1969).
Bennett, B. D. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009).
Mathew, R., Degenhardt, K., Haramaty, L., Karp, C. M. & White, E. Immortalized mouse epithelial cell models to study the role of apoptosis in cancer. Methods Enzymol. 446, 77–106 (2008).
Pisithkul, T., Jacobson, T. B., O’Brien, T. J., Stevenson, D. M. & Amador-Noguez, D. Phenolic amides are potent inhibitors of de novo nucleotide. Appl. Environ. Microbiol. 81, 5761–5772 (2015).
Clasquin, M. F., Melamud, E. & Rabinowitz, J. D. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics 37, 14 11.1–14.11.23 (2012).
Su, X., Lu, W. & Rabinowitz, J. D. Metabolite spectral accuracy on orbitraps. Anal. Chem. 89, 5940–5948 (2017).
Fan, J. et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol. Syst. Biol. 9, 712 (2013).
Carrieri, D. et al. Identification and quantification of water-soluble metabolites by cryoprobe-assisted nuclear magnetic resonance spectroscopy applied to microbial fermentation. Magn. Reson. Chem. 47, S138–S146 (2009).
Hwang, T. L. & Shaka, A. J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. A 112, 275–279 (1995).
Antoniewicz, M. R., Kelleher, J. K. & Stephanopoulos, G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab. Eng. 8, 324–337 (2006).
Acknowledgements
This work was supported by a Department of Energy (DOE) grant (no. DE-SC0012461 to J.D.R.), the Center for Advanced Bioenergy and Bioproducts Innovation (grant no. DE-SC0018420, subcontract to J.D.R.), the Center for Bioenergy Innovation (grant no. DE-AC05-00OR22725, subcontract to D.A.-N.) and ExxonMobil through its membership in the Princeton E-ffiliates Partnership of the Andlinger Center for Energy and the Environment. The Center for Advanced Bioenergy and Bioproducts Innovation and the Center for Bioenergy Innovation are both U.S. Department of Energy Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the DOE Office of Science. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the U.S. Department of Energy.
Author information
Authors and Affiliations
Contributions
J.O.P., L.B.T., D.A.-N. and J.D.R. designed the study. J.O.P., D.B.K., T.B.J., S.H.-J.L. and M.B.H. carried out the E. coli experiments. J.O.P., L.B.T. and M.H.W. carried out the mammalian cell experiments. Z.Z., D.M.S. and D.A.-N. carried out the Clostridia experiments. J.O.P., M.H.W. and S.A.R. developed the computational tools for reaction flux, reversibility and Gibbs free energy quantitation. J.O.P., D.A.-N. and J.D.R. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–15, Supplementary Tables 1–13 and Supplementary Notes.
Rights and permissions
About this article
Cite this article
Park, J.O., Tanner, L.B., Wei, M.H. et al. Near-equilibrium glycolysis supports metabolic homeostasis and energy yield. Nat Chem Biol 15, 1001–1008 (2019). https://doi.org/10.1038/s41589-019-0364-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0364-9
This article is cited by
-
Systems engineering of Escherichia coli for high-level glutarate production from glucose
Nature Communications (2024)
-
A parallel glycolysis provides a selective advantage through rapid growth acceleration
Nature Chemical Biology (2024)
-
Mitochondrial ATP generation is more proteome efficient than glycolysis
Nature Chemical Biology (2024)
-
Thermodynamic principle to enhance enzymatic activity using the substrate affinity
Nature Communications (2023)
-
Role of green financing and financial inclusion to develop the cleaner environment for macroeconomic stability: Inter-temporal analysis of ASEAN economies
Economic Change and Restructuring (2023)