Abstract
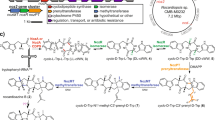
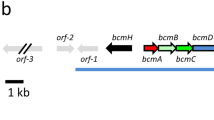
Fosmidomycin and related molecules comprise a family of phosphonate natural products with potent antibacterial, antimalarial and herbicidal activities. To understand the biosynthesis of these compounds, we characterized the fosmidomycin producer, Streptomyces lavendulae, using biochemical and genetic approaches. We were unable to elicit production of fosmidomycin, instead observing the unsaturated derivative dehydrofosmidomycin, which we showed potently inhibits 1-deoxy-d-xylulose-5-phosphate reductoisomerase and has bioactivity against a number of bacteria. The genes required for dehydrofosmidomycin biosynthesis were established by heterologous expression experiments. Bioinformatics analyses, characterization of intermediates and in vitro biochemistry show that the biosynthetic pathway involves conversion of a two-carbon phosphonate precursor into the unsaturated three-carbon product via a highly unusual rearrangement reaction, catalyzed by the 2-oxoglutarate dependent dioxygenase DfmD. The required genes and biosynthetic pathway for dehydrofosmidomycin differ substantially from that of the related natural product FR-900098, suggesting that the ability to produce these bioactive molecules arose via convergent evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that all relevant data supporting the findings of this study are available within the paper and its Supplementary Information.
References
Antibiotic Resistance Threats in the United States, Antibiotic/Antimicrobial Resistance (The Centers for Disease Control and Prevention, 2013).
World Malaria Report 2016 (World Health Organization, 2016).
Bain, C., Selfa, T., Dandachi, T. & Velardi, S. ‘Superweeds’ or ‘survivors’? Framing the problem of glyphosate resistant weeds and genetically engineered crops. J. Rural Stud. 51, 211–221 (2017).
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 (2016).
Doroghazi, J. R. et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968 (2014).
Ju, K.-S. et al. Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc. Natl Acad. Sci. USA 112, 12175–12180 (2015).
Iguchi, E., Okuhara, M., Kohsaka, M., Aoki, H. & Imanaka, H. Studies on new phosphonic acid antibiotics. II. Taxonomic studies on producing organisms of the phosphonic acid and related compounds. J. Antibiot. 33, 19–23 (1980).
Okuhara, M. et al. Studies on new phosphonic acid antibiotics. III. Isolation and characterization of FR-31564, FR-32863 and FR-33289. J. Antibiot. 33, 24–28 (1980).
Kuroda, Y. et al. Studies on new phosphonic acid antibiotics. IV. Structure determination of FR-33289, FR-31564 and FR-32863. J. Antibiot. 33, 29–35 (1980).
Okuhara, M. et al. Studies on new phosphonic acid antibiotics. I. FR-900098, isolation and characterization. J. Antibiot. 33, 13–17 (1980).
Davey, M. S. et al. A promising target for treatment of multidrug-resistant bacterial infections. Antimicrob. Agents Chemother. 55, 3635–3636 (2011).
Jomaa, H. et al. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285, 1573–1576 (1999).
Fernandes, J. F. et al. Fosmidomycin as an antimalarial drug: a meta-analysis of clinical trials. Future Microbiol. 10, 1375–1390 (2015).
Kuzuyama, T., Shimizu, T., Takahashi, S. & Seto, H. Fosmidomycin, a specific inhibitor of 1-deoxy-d-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett. 39, 7913–7916 (1998).
Coppens, I. Targeting lipid biosynthesis and salvage in apicomplexan parasites for improved chemotherapies. Nat. Rev. Microbiol. 11, 823–835 (2013).
Hartmann, M. et al. The effect of MEP pathway and other inhibitors on the intracellular localization of a plasma membrane-targeted, isoprenylable GFP reporter protein in tobacco BY-2 cells. F1000Res. 2, 170 (2013).
Hunter, W. N. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 282, 21573–21577 (2007).
Lichtenthaler, H. K., Zeidler, J., Schwender, J. & Müller, C. The non-mevalonate isoprenoid biosynthesis of plants as a test system for new herbicides and drugs against pathogenic bacteria and the malaria parasite. Z. Naturforsch. C 55, 305–313 (2000).
Eliot, A. C. et al. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem. Biol. 15, 765–770 (2008).
Johannes, T. W. et al. Deciphering the late biosynthetic steps of antimalarial compound FR-900098. Chem. Biol. 17, 57–64 (2010).
Peck, S. C. & van der Donk, W. A. Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology. Curr. Opin. Chem. Biol. 17, 580–588 (2013).
Metcalf, W. W. & van der Donk, W. A. Biosynthesis of phosphonic and phosphinic acid natural products. Annu. Rev. Biochem. 78, 65–94 (2009).
Horsman, G. P. & Zechel, D. L. Phosphonate biochemistry. Chem. Rev. 117, 5704–5783 (2017).
Sindt, M., Stephan, B., Schneider, M. & Mieloszynski, J. L. Chemical shift prediction of 31 P-NMR shifts for dialkyl or diaryl phosphonates. Phosphorus Sulfur Silicon Relat. Elem. 174, 163–175 (2001).
Hemmi, K., Takeno, H., Hashimoto, M. & Kamiya, T. Studies on phosphonic acid antibiotics. IV. Synthesis and antibacterial activity of analogs of 3-(N-acetyl-N-hydroxyamino)-propylphosphonic acid (FR-900098). Chem. Pharm. Bull. 30, 111–118 (1982).
Wang, X. et al. MEPicides: α,β-unsaturated fosmidomycin analogues as DXR Inhibitors against malaria. J. Med. Chem. 61, 8847–8858 (2018).
Waditee, R. et al. Isolation and functional characterization of N-methyltransferases that catalyze betaine synthesis from glycine in a halotolerant photosynthetic organism Aphanothece halophytica. J Biol. Chem. 278, 4932–4942 (2003).
Martinez-Morales, F., Schobert, M., López-Lara, I. M. & Geiger, O. Pathways for phosphatidylcholine biosynthesis in bacteria. Microbiology 149, 3461–3471 (2003).
Geiger, O., López-Lara, I. M. & Sohlenkamp, C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim. Biophys. Acta – Mol. Cell Biol. Lipids 1831, 503–513 (2013).
Blodgett, J. A. V. et al. Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nat. Chem. Biol. 3, 480–485 (2007).
Loenarz, C. & Schofield, C. J. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem. Sci. 36, 7–18 (2011).
Herr, C. Q. & Hausinger, R. P. Amazing diversity in biochemical roles of Fe(II)/2-oxoglutarate oxygenases. Trends Biochem. Sci. 43, 517–532 (2018).
Islam, M. S., Leissing, T. M., Chowdhury, R., Hopkinson, R. J. & Schofield, C. J. 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem. 87, 585–620 (2018).
Dong, C., Zhang, H., Xu, C., Arrowsmith, C. H. & Min, J. Structure and function of dioxygenases in histone demethylation and DNA/RNA demethylation. IUCr. J. 1, 540–549 (2014).
Valegård, K. et al. Structure of a cephalosporin synthase. Nature 394, 805–809 (1998).
Salowe, S. P., Marsh, E. N. & Townsend, C. A. Purification and characterization of clavaminate synthase from Streptomyces clavuligerus: an unusual oxidative enzyme in natural product biosynthesis. Biochemistry 29, 6499–6508 (1990).
Zhang, Z. et al. Crystal structure of a clavaminate synthase-Fe(II)-2-oxoglutarate-substrate-NO complex: evidence for metal centered rearrangements. FEBS Lett. 517, 7–12 (2002).
Rydzik, A. M. et al. Comparison of the substrate selectivity and biochemical properties of human and bacterial γ-butyrobetaine hydroxylase. Org. Biomol. Chem. 12, 6354–6358 (2014).
Henry, L., Leung, I. K. H., Claridge, T. D. W. & Schofield, C. J. γ-Butyrobetaine hydroxylase catalyses a Stevens type rearrangement. Bioorg. Med. Chem. Lett. 22, 4975–4978 (2012).
Leung, I. K. H. et al. Structural and mechanistic studies on γ-utyrobetaine hydroxylase. Chem. Biol. 17, 1316–1324 (2010).
Kobayashi, S., Kuzuyama, T. & Seto, H. Characterization of the fomA and fomB gene products from Streptomyces wedmorensis, which confer fosfomycin resistance on Escherichia coli. Antimicrob. Agents Chemother. 44, 647–650 (2000).
Kim, S. Y. et al. Different biosynthetic pathways to fosfomycin in Pseudomonas syringae and Streptomyces peciess. Antimicrob. Agents Chemother. 56, 4175–4183 (2012).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Patel, J. B. et al. Methods for Dilution Antimicrobial Susceptibilities Tests for Bacteria tThat Grow Aerobically; Approved Standard 10th edn. (Clinical and Laboratory Standards Institute, 2015).
Blodgett, J. A. V., Zhang, J. K. & Metcalf, W. W. Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob. Agents Chemother. 49, 230–240 (2005).
Yu, X., Price, N. P. J., Evans, B. S. & Metcalf, W. W. Purification and characterization of phosphonoglycans from Glycomyces sp. strain NRRL B-16210 and Stackebrandtia nassauensis NRRL B-16338. J. Bacteriol. 196, 1768–1779 (2014).
Wang, S., Cui, X. & Fang, G. Rapid determination of formaldehyde and sulfur dioxide in food products and Chinese herbals. Food Chem. 103, 1487–1493 (2007).
Acknowledgements
We thank M. Goettge and K. Wang for helpful discussions, X. Guan for his assistance with NMR experiments, Z. Li for his assistance with HRMS experiments, the UIUC Core Sequencing Facility for sequencing and P. Hergenrother (Chemistry Department, UIUC) for providing strains. This work was funded by the National Institutes of Health (grant nos. P01 GM077596 and R01GM127659 to W.W.M. and F32GM120999 to E.I.P.). NMR spectra were recorded on an instrument purchased with support from NIH grant no. S10 RR028833.
Author information
Authors and Affiliations
Contributions
E.I.P., A.E. and K.-S.J. optimized growth conditions. E.I.P. performed the chemical syntheses, bioactivity analyses and in vitro characterization of enzymes. A.C.E. made the heterologous expression vector. E.I.P. performed the heterologous expression studies and the deletion mutant studies. E.I.P. and W.W.M. analyzed the data and wrote the manuscript, with assistance of A.E., A.C.E. and K.-S.J.
Corresponding author
Ethics declarations
Competing interests
W.W.M. has financial interest in Microbial Pharmaceuticals.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–5, Supplementary Figures 1–14 and Supplementary Note 1
Rights and permissions
About this article
Cite this article
Parkinson, E.I., Erb, A., Eliot, A.C. et al. Fosmidomycin biosynthesis diverges from related phosphonate natural products. Nat Chem Biol 15, 1049–1056 (2019). https://doi.org/10.1038/s41589-019-0343-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0343-1
This article is cited by
-
Chiral phosphoric acid-catalyzed enantioselective phosphinylation of 3,4-dihydroisoquinolines with diarylphosphine oxides
Communications Chemistry (2023)