Abstract
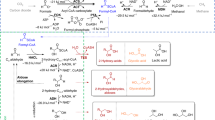
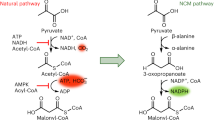
Despite the potential of biotechnological processes for one-carbon (C1) bioconversion, efficient biocatalysts required for their implementation are yet to be developed. To address intrinsic limitations of native C1 biocatalysts, here we report that 2-hydroxyacyl CoA lyase (HACL), an enzyme involved in mammalian α-oxidation, catalyzes the ligation of carbonyl-containing molecules of different chain lengths with formyl-coenzyme A (CoA) to produce C1-elongated 2-hydroxyacyl-CoAs. We discovered and characterized a prokaryotic variant of HACL and identified critical residues for this newfound activity, including those supporting the hypothesized thiamine pyrophosphate-dependent acyloin condensation mechanism. The use of formyl-CoA as a C1 donor provides kinetic advantages and enables C1 bioconversion to multi-carbon products, demonstrated here by engineering an Escherichia coli whole-cell biotransformation system for the synthesis of glycolate and 2-hydroxyisobutyrate from formaldehyde and formaldehyde plus acetone, respectively. Our work establishes a new approach for C1 bioconversion and the potential for HACL-based pathways to support synthetic methylotrophy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data supporting the findings of this study are included in the paper and its supplementary information files.
References
Peralta-Yahya, P. P., Zhang, F., del Cardayre, S. B. & Keasling, J. D. Microbial engineering for the production of advanced biofuels. Nature 488, 320–328 (2012).
Casini, A. et al. A pressure test to make 10 molecules in 90 days: external evaluation of methods to engineer biology. J. Am. Chem. Soc. 140, 4302–4316 (2018).
Claassens, N. J., Sousa, D. Z., Dos Santos, V. A. P. M., De Vos, W. M. & Van Der Oost, J. Harnessing the power of microbial autotrophy. Nat. Rev. Microbiol. 14, 692–706 (2016).
Bennett, R. K., Steinberg, L. M., Chen, W. & Papoutsakis, E. T. Engineering the bioconversion of methane and methanol to fuels and chemicals in native and synthetic methylotrophs. Curr. Opin. Biotechnol. 50, 81–93 (2018).
Antonovsky, N., Gleizer, S. & Milo, R. Engineering carbon fixation in E. coli: from heterologous RuBisCO expression to the Calvin–Benson–Bassham cycle. Curr. Opin. Biotechnol. 47, 83–91 (2017).
Clomburg, J. M., Crumbley, A. M. & Gonzalez, R. Industrial biomanufacturing: the future of chemical production. Science 355, 1–10 (2017).
Casteels, M., Foulon, V., Mannaerts, G. P. & Van Veldhoven, P. P. Alpha-oxidation of 3-methyl-substituted fatty acids and its thiamine dependence. Eur. J. Biochem. 270, 1619–1627 (2003).
Foulon, V. et al. Breakdown of 2-hydroxylated straight chain fatty acids via peroxisomal 2-hydroxyphytanoyl-CoA lyase: a revised pathway for the alpha-oxidation of straight chain fatty acids. J. Biol. Chem. 280, 9802–9812 (2005).
Foulon, V. et al. Purification, molecular cloning, and expression of 2-hydroxyphytanoyl-CoA lyase, a peroxisomal thiamine pyrophosphate-dependent enzyme that catalyzes the carbon-carbon bond cleavage during alpha-oxidation of 3-methyl-branched fatty acids. Proc. Natl Acad. Sci. USA 96, 10039–10044 (1999).
Müller, M. & Sprenger, G. A. in Thiamine: Catalytic Mechanisms in Normal and Disease States (eds Jordan, F. & Patel, M. S.) 82–84 (CRC Press, 2003).
Dellomonaco, C., Clomburg, J. M., Miller, E. N. & Gonzalez, R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 476, 355–359 (2011).
Cheong, S., Clomburg, J. M. & Gonzalez, R. Energy-and carbon-efficient synthesis of functionalized small molecules in bacteria using non-decarboxylative Claisen condensation reactions. Nat. Biotechnol. 34, 556–561 (2016).
Jankowski, M. D., Henry, C. S., Broadbelt, L. J. & Hatzimanikatis, V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys. J. 95, 1487–1499 (2008).
Caspi, R. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 42, D459–D471 (2014).
Jansen, G. A. & Wanders, R. J. A. Alpha-oxidation. Biochim. Biophys. Acta 1763, 1403–1412 (2006).
Croes, K., Casteels, M., Dieuaide-Noubhani, M., Mannaerts, G. P. & Van Veldhoven, P. P. Stereochemistry of the alpha-oxidation of 3-methyl-branched fatty acids in rat liver. J. Lipid Res. 40, 601–609 (1999).
Ring, M. W., Schwär, G. & Bode, H. B. Biosynthesis of 2-hydroxy and iso-even fatty acids is connected to sphingolipid formation in myxobacteria. Chem. Bio. Chem. 10, 2003–2010 (2009).
Landry, Z., Swan, B. K., Herndl, G. J., Stepanauskas, R. & Giovannoni, S. J. SAR202 genomes from the dark ocean predict pathways for the oxidation of recalcitrant dissolved organic matter. mBio. 8, e00413–e00417 (2017).
Madden, T. T. The BLAST sequence analysis tool. NCBI Handbook 2nd edn, 1–12 (eds. Hoeppner, M. and Ostell, J.; National Library of Medicine, 2013); https://www.unmc.edu/bsbc/docs/NCBI_blast.pdf
Lung, H. Y., Baetz, A. L. & Peck, A. B. Molecular cloning, DNA sequence, and gene expression of the oxalyl-coenzyme A decarboxylase gene, oxc, from the bacterium Oxalobacter formigenes. J. Bacteriol. 176, 2468–2472 (1994).
Bar-Even, A. et al. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410 (2011).
Schwander, T., von Borzyskowski, S., Burgener, L., Cortina, S. & Erb, N. S. A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354, 900–904 (2016).
Siegel, J. B. et al. Computational protein design enables a novel one-carbon assimilation pathway. Proc. Natl Acad. Sci. USA 112, 3704–3709 (2015).
Choi, Y. J. et al. Novel, versatile, and tightly regulated expression system for Escherichia coli strains. Appl. Environ. Microbiol. 76, 5058–5066 (2010).
Rohwerder, T. & Müller, R. H. Biosynthesis of 2-hydroxyisobutyric acid (2-HIBA) from renewable carbon. Microb. Cell Fact. 9, 1–10 (2010).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, gky427 (2018).
Berthold, C. L. et al. Crystallographic snapshots of oxalyl-CoA decarboxylase give insights into catalysis by nonoxidative ThDP-dependent decarboxylases. Structure 15, 853–861 (2007).
Sly, W. & Stadtman, E. Formyl coenzyme A, an intermediate in the formate-dependent decomposition of acetyl phosphate in Clostridium kluyveri. J. Biol. Chem. 238, 2632–2638 (1963).
Müller, J. E. N. et al. Engineering Escherichia coli for methanol conversion. Metab. Eng. 28, 190–201 (2015).
Bahmanpour, A. M., Hoadley, A. & Tanksale, A. Critical review and exergy analysis of formaldehyde production processes. Rev. Chem. Eng. 30, 583–604 (2014).
Heim, L. E., Konnerth, H. & Prechtl, M. H. G. Future perspectives for formaldehyde: pathways for reductive synthesis and energy storage. Green Chem. 19, 2347–2355 (2017).
Pereira, B. et al. Efficient utilization of pentoses for bioproduction of the renewable two-carbon compounds ethylene glycol and glycolate. Metab. Eng. 34, 80–87 (2016).
Koivistoinen, O. M. et al. Glycolic acid production in the engineered yeasts Saccharomyces cerevisiae and Kluyveromyces lactis. Microb. Cell Fact. 12, 82 (2013).
Krakow, G., Barkulis, S. S. & Hayashi, J. A. Glyoxylic acid carboligase: an enzyme present in glycolate-grown Escherichia coli. J. Bacteriol. 81, 509–518 (1961).
Miller, J. H. Experiments in Molecular Genetics (Cold Spring Harbor Laboratory Press, 1972).
Jiang, Y. et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514 (2015).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Fong, J. C. & Schulz, H. Short-chain and long-chain enoyl-CoA hydratases from pig heart muscle. Methods Enzymol. 71, 390–398 (1981).
Parasaran, T. & Tarbell, D. S. Formic ethylcarbonic anhydride. J. Org. Chem. 29, 3422–3423 (1964).
Taylor, D. C., Weber, N. & MacKenzie, S. L. in CRC Handbook of Chromatography. Analysis of Lipids (eds Mukherjee, K. D. & Weber, N.) (CRC Press, 1993).
Severn, D. J., Johnson, M. E. & Olson, N. F. Determination of lactic acid in cheddar cheese and calcium lactate crystals. J. Dairy Sci. 69, 2027–2030 (2010).
Neidhardt, F. C., Bloch, P. L. & Smith, D. F. Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 (1974).
Papadopoulos, J. S. & Agarwala, R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 (2007).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank S. Garg for assistance with protein purification methods and S. Cheong for assistance with genetic methods. This work was partially supported by a grant from the US National Science Foundation (no. CBET-1605999).
Author information
Authors and Affiliations
Contributions
R.G. conceptualized the research and supervised the project. A.C., J.M.C., S.Q. and R.G. designed the methodology. A.C. identified and characterized HACL variants and performed cell-free experiments. S.Q. modeled and analyzed the structure of RuHACL and designed mutations. J.M.C. performed whole-cell experiments. A.C. and S.Q. constructed E. coli strains. A.C., J.M.C. and S.Q. analyzed the data. A.C. and R.G. prepared the manuscript with feedback from all authors.
Corresponding author
Ethics declarations
Competing interests
A.C., J.M.C. and R.G. are co-inventors and assignees on a patent application (PCT/US2015/058121), which relates to the reported research. Correspondence and requests for materials should be addressed to ramongonzale@usf.edu.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1–3, Supplementary Figs. 1–11
Supplementary Dataset
RuHACLmodel
Rights and permissions
About this article
Cite this article
Chou, A., Clomburg, J.M., Qian, S. et al. 2-Hydroxyacyl-CoA lyase catalyzes acyloin condensation for one-carbon bioconversion. Nat Chem Biol 15, 900–906 (2019). https://doi.org/10.1038/s41589-019-0328-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0328-0
This article is cited by
-
Engineering a synthetic energy-efficient formaldehyde assimilation cycle in Escherichia coli
Nature Communications (2023)
-
Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo
Nature Communications (2023)
-
Physiological limitations and opportunities in microbial metabolic engineering
Nature Reviews Microbiology (2022)
-
A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nature Catalysis (2021)
-
A new path for one-carbon conversion
Nature Metabolism (2021)