Abstract
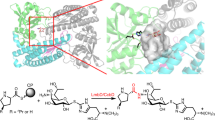
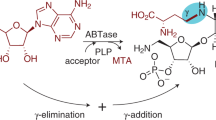
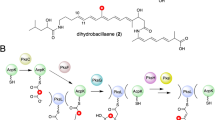
Glycosylation is a common modification reaction in natural product biosynthesis and has been known to be a post-assembly line tailoring process in glycosylated polyketide biosynthesis. Here, we show that in pactamycin biosynthesis, glycosylation can take place on an acyl carrier protein (ACP)-bound polyketide intermediate. Using in vivo gene inactivation, chemical complementation and in vitro pathway reconstitution, we demonstrate that the 3-aminoacetophenone moiety of pactamycin is derived from 3-aminobenzoic acid by a set of discrete polyketide synthase proteins via a 3-(3-aminophenyl)3-oxopropionyl-ACP intermediate. This ACP-bound intermediate is then glycosylated by an N-glycosyltransferase, PtmJ, providing a sugar precursor for the formation of the aminocyclopentitol core structure of pactamycin. This is the first example of glycosylation of a small molecule while tethered to a carrier protein. Additionally, we demonstrate that PtmO is a hydrolase that is responsible for the release of the ACP-bound product to a free β-ketoacid that subsequently undergoes decarboxylation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data that support the conclusions are included in the published paper and its Supplementary Information, or are available from the authors on request.
References
Mao, Y., Varoglu, M. & Sherman, D. H. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem. Biol. 6, 251–263 (1999).
Kudo, F., Kasama, Y., Hirayama, T. & Eguchi, T. Cloning of the pactamycin biosynthetic gene cluster and characterization of a crucial glycosyltransferase prior to a unique cyclopentane ring formation. J. Antibiot. 60, 492–503 (2007).
Ito, T. et al. Deciphering pactamycin biosynthesis and engineered production of new pactamycin analogues. Chem. Bio. Chem. 10, 2253–2265 (2009).
Rinehart, K. L. Jr., Weller, D. D. & Pearce, C. J. Recent biosynthetic studies on antibiotics. J. Nat. Prod. 43, 1–20 (1980).
Almabruk, K. H. et al. Mutasynthesis of fluorinated pactamycin analogues and their antimalarial activity. Org. Lett. 15, 1678–1681 (2013).
Hirayama, A., Eguchi, T. & Kudo, F. A single PLP-dependent enzyme PctV catalyzes the transformation of 3-dehydroshikimate into 3-aminobenzoate in the biosynthesis of pactamycin. Chem. Bio. Chem. 14, 1198–1203 (2013).
Rinehart, K. L. Jr. Biosynthesis and mutasynthesis of aminocyclitol antibiotics. Jpn J. Antibiot. 32, S32–S46 (1979).
Hirayama, A., Miyanaga, A., Kudo, F. & Eguchi, T. Mechanism-based trapping of the quinonoid intermediate by using the K276R mutant of PLP-dependent 3-aminobenzoate synthase PctV in the biosynthesis of pactamycin. Chem. Bio. Chem. 16, 2484–2490 (2015).
Abugrain, M. E. et al. Interrogating the tailoring steps of pactamycin biosynthesis and accessing new pactamycin analogues. Chem. Bio. Chem. 17, 1585–1588 (2016).
Abugrain, M. E., Brumsted, C. J., Osborn, A. R., Philmus, B. & Mahmud, T. A highly promiscuous β-ketoacyl-ACP synthase (KAS) III-like protein is involved in pactamycin biosynthesis. ACS Chem. Biol. 12, 362–366 (2017).
Bibb, M. J., Sherman, D. H., Omura, S. & Hopwood, D. A. Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin. Gene 142, 31–39 (1994).
Ahlert, J. et al. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 297, 1173–1176 (2002).
Mao, Y., Varoglu, M. & Sherman, D. H. Genetic localization and molecular characterization of two key genes (mitAB) required for biosynthesis of the antitumor antibiotic mitomycin C. J. Bacteriol. 181, 2199–2208 (1999).
Lu, W., Roongsawang, N. & Mahmud, T. Biosynthetic studies and genetic engineering of pactamycin analogs with improved selectivity toward malarial parasites. Chem. Biol. 18, 425–431 (2011).
Upson, R. H., Haugland, R. P., Malekzadeh, M. N. & Haugland, R. P. A spectrophotometric method to measure enzymatic activity in reactions that generate inorganic pyrophosphate. Anal. Biochem. 243, 41–45 (1996).
Yang, J. et al. Nucleotidylation of unsaturated carbasugar in validamycin biosynthesis. Org. Biomol. Chem. 9, 438–449 (2011).
Jia, X. Y. et al. Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem. Biol. 13, 575–585 (2006).
Mondal, S., Hsiao, K. & Goueli, S. A. Utility of adenosine monophosphate detection system for monitoring the activities of diverse enzyme reactions. Assay Drug Dev. Technol. 15, 330–341 (2017).
Asamizu, S., Yang, J., Almabruk, K. H. & Mahmud, T. Pseudoglycosyltransferase catalyzes nonglycosidic C–N coupling in validamycin a biosynthesis. J. Am. Chem. Soc. 133, 12124–12135 (2011).
Adams, E. S. & Rinehart, K. L. Directed biosynthesis of 5”-fluoropactamycin in Streptomyces pactum. J. Antibiot. 47, 1456–1465 (1994).
Schmelz, S. & Naismith, J. H. Adenylate-forming enzymes. Curr. Opin. Struct. Biol. 19, 666–671 (2009).
August, P. R. et al. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem. Biol. 5, 69–79 (1998).
Yu, T. W. et al. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl Acad. Sci. USA 99, 7968–7973 (2002).
Admiraal, S. J., Khosla, C. & Walsh, C. T. A Switch for the transfer of substrate between nonribosomal peptide and polyketide modules of the rifamycin synthetase assembly line. J. Am. Chem. Soc. 125, 13664–13665 (2003).
Zawada, R. J. & Khosla, C. Domain analysis of the molecular recognition features of aromatic polyketide synthase subunits. J. Biol. Chem. 272, 16184–16188 (1997).
Sherman, D. H., Kim, E. S., Bibb, M. J. & Hopwood, D. A. Functional replacement of genes for individual polyketide synthase components in Streptomyces coelicolor A3(2) by heterologous genes from a different polyketide pathway. J. Bacteriol. 174, 6184–6190 (1992).
Lu, W., Alanzi, A. R., Abugrain, M. E., Ito, T. & Mahmud, T. Global and pathway-specific transcriptional regulations of pactamycin biosynthesis in Streptomyces pactum. Appl. Microbiol. Biotechnol. 102, 10589–10601 (2018).
Hirayama, A., Chu, J., Goto, E., Kudo, F. & Eguchi, T. NAD+-dependent dehydrogenase PctP and pyridoxal 5´-phosphate dependent aminotransferase PctC catalyze the first postglycosylation modification of the sugar intermediate in pactamycin biosynthesis. Chem. Bio. Chem. 19, 126–130 (2018).
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (The John Innes Foundation, 2000).
Green, M. R. & Sambrook, J. in A Laboratory Manual (eds Inglis, J., Boyle, A. & Gann, A.) Ch 3 (Cold Spring Harbor Laboratory Press, 2012).
Ishikawa, J. & Hotta, K. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174, 251–253 (1999).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
He, Y. et al. Two pHZ1358-derivative vectors for efficient gene knockout in Streptomyces. J. Microbiol. Biotechnol. 20, 678–682 (2010).
Acknowledgements
The authors thank M. Zabriskie and B. Philmus for critical reading of this manuscript, W. Lu for performing some preliminary work, L. Yang and J. Morre for providing assistance in protein mass spectrometry analysis and A. DeBarber for high-resolution mass spectrometry measurements. This work was supported by grant nos. GM112068 (to T.M.) and AI129957 (to T.M.) from the National Institute of General Medical Sciences and the National Institute of Allergy and Infectious Diseases, respectively. The content is solely the responsibility of the authors and does not represent the official views of the National Institute of General Medical Sciences, the National Institute of Allergy and Infectious Diseases or the National Institutes of Health (NIH).
Author information
Authors and Affiliations
Contributions
A.A.E. designed and performed the enzymatic assays and analyzed the data; M.E.A. designed and performed the gene inactivation and complementation experiments and analyzed the data; C.J.B. designed and performed the chemical synthesis; T.M. designed the overall project, analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information
Supplementary Tables 1–6 and Supplementary Figures 1–22
Supplementary Note
Synthetic Procedures
Rights and permissions
About this article
Cite this article
Eida, A.A., Abugrain, M.E., Brumsted, C.J. et al. Glycosylation of acyl carrier protein-bound polyketides during pactamycin biosynthesis. Nat Chem Biol 15, 795–802 (2019). https://doi.org/10.1038/s41589-019-0314-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0314-6