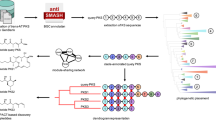
Abstract
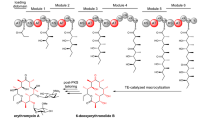
Bacterial trans-acyltransferase polyketide synthases (trans-AT PKSs) are among the most complex known enzymes from secondary metabolism and are responsible for the biosynthesis of highly diverse bioactive polyketides. However, most of these metabolites remain uncharacterized, since trans-AT PKSs frequently occur in poorly studied microbes and feature a remarkable array of non-canonical biosynthetic components with poorly understood functions. As a consequence, genome-guided natural product identification has been challenging. To enable de novo structural predictions for trans-AT PKS-derived polyketides, we developed the trans-AT PKS polyketide predictor (TransATor). TransATor is a versatile bio- and chemoinformatics web application that suggests informative chemical structures for even highly aberrant trans-AT PKS biosynthetic gene clusters, thus permitting hypothesis-based, targeted biotechnological discovery and biosynthetic studies. We demonstrate the applicative scope in several examples, including the characterization of new variants of bioactive natural products as well as structurally new polyketides from unusual bacterial sources.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
GenBank: the Aquimarina sp. Aq349 and Aq78 genome sequence harboring the cuniculene BGC has been submitted to the European Nucleotide Archive (accession codes OMKB01000001–OMKB01000022 and OMKF01000001–OMKF01000170). The leptolyngbyalide BGC (BK010645) from Leptolyngbyia sp. PCC 7375, and the tartrolon BGC (BK010667) from G. sunshinyii were deposited in GenBank. MIBiG: the lepolyngbyalide (BGC0001835), tartrolon (BGC0001836) and cuniculene (BGC0001855) BGCs were deposited in the MIBiG database. The TransATor web server is freely accessible at http://transator.ethz.ch. Code for TransATor pipeline/web application is available at https://github.com/pcm32/transator-container.
References
Helfrich, E. J. & Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 33, 231–316 (2016).
El-Sayed, A. K. et al. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 10, 419–430 (2003).
Pulsawat, N., Kitani, S. & Nihira, T. Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae. Gene 393, 31–42 (2007).
Ueoka, R., Bortfeld-Miller, M., Morinaka, B. I., Vorholt, J. A. & Piel, J. Toblerols, cyclopropanol-containing modulators of methylobacterial antibiosis generated by an unusual polyketide synthase. Angew. Chem. Int. Ed. Engl. 57, 977–981 (2017).
Möbius, N. et al. Biosynthesis of the respiratory toxin bongkrekic acid in the pathogenic bacterium Burkholderia gladioli. Chem. Biol. 19, 1164–1174 (2012).
Partida-Martinez, L. P. & Hertweck, C. A gene cluster encoding rhizoxin biosynthesis in Burkholderia rhizoxina, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chem. Bio. Chem. 8, 41–45 (2007).
O’Brien, R. V., Davis, R. W., Khosla, C. & Hillenmeyer, M. E. Computational identification and analysis of orphan assembly-line polyketide synthases. J. Antibiot. 67, 89–97 (2014).
Blin, K., Medema, M. H., Kottmann, R., Lee, S. Y. & Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 45, 555–559 (2016).
Liu, L., Hao, T., Xie, Z., Horsman, G. P. & Chen, Y. Genome mining unveils widespread natural product biosynthetic capacity in human oral microbe Streptococcus mutans. Sci. Rep 6, 37479 (2016).
Ueoka, R. et al. Metabolic and evolutionary origin of actin-inhibiting polyketides from diverse organisms. Nat. Chem. Biol. 11, 705–712 (2015).
Nakabachi, A. et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 23, 1478–1484 (2013).
Wilson, M. C. et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014).
Blin, K. et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, 36–41 (2017).
Skinnider, M. A. et al. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 43, 9645–9662 (2015).
Nguyen, T. et al. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 26, 225–233 (2008).
Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48, 4688–4716 (2009).
Helfrich, E. J. N., Reiter, S. & Piel, J. Recent advances in genome-based polyketide discovery. Curr. Opin. Biotechnol. 29, 107–115 (2014).
Bretschneider, T. et al. Vinylogous chain branching catalysed by a dedicated polyketide synthase module. Nature 502, 124–128 (2013).
Pöplau, P., Frank, S., Morinaka, B. I. & Piel, J. An enzymatic domain for the formation of cyclic ethers in complex polyketides. Angew. Chem. Int. Ed. Engl. 52, 13215–13218 (2013).
Jenner, M. et al. Substrate specificity in ketosynthase domains from trans-AT polyketide synthases. Angew. Chem. Int. Ed. Engl. 52, 1143–1147 (2013).
Jenner, M. et al. Acyl-chain elongation drives ketosynthase substrate selectivity in trans-acyltransferase polyketide synthases. Angew. Chem. Int. Ed. Engl. 54, 1817–1821 (2015).
Teta, R. et al. Genome mining reveals trans-AT polyketide synthase directed antibiotic biosynthesis in the bacterial phylum bacteroidetes. Chem. Bio. Chem. 11, 2506–2512 (2010).
Kampa, A. et al. Metagenomic natural product discovery in lichen provides evidence for specialized biosynthetic pathways in diverse symbioses. Proc. Natl Acad. Sci. USA 110, 3129–3127 (2013).
Sundaram, S., Heine, D. & Hertweck, C. Polyketide synthase chimeras reveal key role of ketosynthase domain in chain branching. Nat. Chem. Biol. 11, 949–951 (2015).
Fisch, K. M. et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 5, 494–501 (2009).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, 29–37 (2011).
Röttig, M. et al. NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, 362–367 (2011).
Steinbeck, C. et al. Recent developments of the chemistry development kit (CDK) - an open-source java library for chemo- and bioinformatics. Curr. Pharm. Des. 12, 2111–2120 (2006).
Reddick, J. J., Antolak, S. A. & Raner, G. M. PksS from Bacillus subtilis is a cytochrome P450 involved in bacillaene metabolism. Biochem. Biophys. Res. Commun. 358, 363–367 (2007).
Moldenhauer, J., Chen, X. H., Borriss, R. & Piel, J. Biosynthesis of the antibiotic bacillaene, the product of a giant polyketide synthase complex of the trans-AT family. Angew. Chem. Int. Ed. Engl. 46, 8195–8197 (2007).
Moldenhauer, J. et al. The final steps of bacillaene biosynthesis in Bacillus amyloliquefaciens FZB42: direct evidence for β,γ dehydration by a trans-acyltransferase polyketide synthase. Angew. Chem. Int. Ed. Engl. 49, 1465–1467 (2010).
Kusebauch, B., Busch, B., Scherlach, K., Roth, M. & Hertweck, C. Functionally distinct modules operate two consecutive α,β→β,γ double-bond shifts in the rhizoxin polyketide assembly line. Angew. Chem. Int. Ed. Engl. 49, 1460–1464 (2010).
MarinLit (accessed April 2017); http://pubs.rsc.org/marinlit
Blunt, J. W., Munro, M.H.G. & Laatsch, H. AntiMarin Database (University of Canterbury, 2006, accessed May 2017); https://www.scienceopen.com/document?vid=03a1a98e-434c-4255-a287-5a900f59d024
Elshahawi, S. I. et al. Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc. Natl Acad. Sci. USA 110, 295–304 (2013).
Shih, P. M. et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl Acad. Sci. USA 110, 1053–1058 (2013).
Williamson, R. T., Boulanger, A., Vulpanovici, A., Roberts, M. A. & Gerwick, W. H. Structure and absolute stereochemistry of phormidolide, a new toxic metabolite from the marine cyanobacterium Phormidium sp. J. Org. Chem. 67, 7927–7936 (2002).
Murakami, M., Matsuda, H., Makabe, K. & Yamaguchi, K. Oscillariolide, a novel macrolide from a blue-green-alga Oscillatoria sp. Tetrahedron Lett. 32, 2391–2394 (1991).
Bertin, M. J. et al. The phormidolide biosynthetic gene cluster: A trans-AT PKS pathway encoding a toxic macrocyclic polyketide. Chem. Bio. Chem. 17, 164–173 (2016).
Esteves, A. I. S., Hardoim, C. C. P., Xavier, J. R., Goncalves, J. M. S. & Costa, R. Molecular richness and biotechnological potential of bacteria cultured from Irciniidae sponges in the north-east Atlantic. FEMS Microbiol. Ecol. 85, 519–536 (2013).
Ma, M., Lohman, J. R., Liu, T. & Shen, B. C-S bond cleavage by a polyketide synthase domain. Proc. Natl Acad. Sci. USA 112, 10359–10364 (2015).
Mast, Y. & Wohlleben, W. Streptogramins: two are better than one! Int. J. Med. Microbiol. 304, 44–50 (2014).
Sudek, S. et al. Identification of the putative bryostatin polyketide synthase gene cluster from ‘Candidatus Endobugula sertula’, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 70, 67–74 (2007).
Eustaquio, A. S., Janso, J. E., Ratnayake, A. S., O’Donnell, C. J. & Koehn, F. E. Spliceostatin hemiketal biosynthesis in Burkholderia spp. is catalyzed by an iron/α-ketoglutarate-dependent dioxygenase. Proc. Natl Acad. Sci. USA 111, 3376–3385 (2014).
Biggins, J. B., Ternei, M. A. & Brady, S. F. Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J. Am. Chem. Soc. 134, 13192–13195 (2012).
Piel, J., Hofer, I. & Hui, D. Evidence for a symbiosis island involved in horizontal acquisition of pederin biosynthetic capabilities by the bacterial symbiont of Paederus fuscipes beetles. J. Bacteriol. 186, 1280–1286 (2004).
Cociancich, S. et al. The gyrase inhibitor albicidin consists of p-aminobenzoic acids and cyanoalanine. Nat. Chem. Biol. 11, 195–197 (2015).
Loper, J. E. et al. Rhizoxin analogs, orfamide A and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environ. Microbiol. 11, 3509–3521 (2016).
Schneider-Poetsch, T. et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 (2010).
Donia, M. S. et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 (2014).
Blin, K., Medema, M. H., Kottmann, R., Lee, S. Y. & Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 45, 555–559 (2017).
Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 27, 996–1047 (2010).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Li, Y. F. et al. Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genet. Biol. 89, 18–28 (2016).
Duan, L., Xu, L., Guo, F., Lee, J. & Yan, B. P. A local-density based spatial clustering algorithm with noise. Inform. Syst. 32, 978–986 (2007).
Yeats, C., Redfern, O. C. & Orengo, C. A fast and automated solution for accurately resolving protein domain architectures. Bioinformatics 26, 745–751 (2010).
Esteves, A. I., Hardoim, C. C., Xavier, J. R., Goncalves, J. M. & Costa, R. Molecular richness and biotechnological potential of bacteria cultured from Irciniidae sponges in the north-east Atlantic. FEMS Microbiol. Ecol. 85, 519–536 (2013).
Overbeek, R. et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214 (2014).
Miyazaki, M., Nagano, Y., Fujiwara, Y., Hatada, Y. & Nogi, Y. Aquimarina macrocephali sp. nov., isolated from sediment adjacent to sperm whale carcasses. Int. J. Syst. Evol. Microbiol. 60, 2298–2302 (2010).
Chung, E. J., Park, J. A., Jeon, C. O. & Chung, Y. R. Gynuella sunshinyii gen. nov., sp. nov., an antifungal rhizobacterium isolated from a halophyte, Carex scabrifolia Steud. Int. J. Syst. Evol. Microbiol. 65, 1038–1043 (2015).
Acknowledgements
J.P. acknowledges funding by the ERC (ERC Advanced Project SynPlex), the SNF (NRP 72 ‘Antimicrobial resistance’, no. 407240_167051) and the DFG Research Unit 854. R.C. acknowledges funding by the Portuguese Foundation for Science and Technology (FCT) through research grants (nos. PTDC/BIA-MIC/3865/2012 and PTDC/MAR-BIO/1547/2014). M.G. thanks the Institut Pasteur funding for the Action Ciblée-Collection. C.S. and P.M. acknowledge generous core funding by the European Molecular Biology Laboratory—European Bioinformatics Institute.
Author information
Authors and Affiliations
Contributions
E.J.N.H., C.S., P.M. and J.P. designed the research. E.J.N.H., M.R., A.B., R.A.M. and J.P. performed bioinformatic analyses. E.J.N.H. performed statistical analyses, compiled the training dataset for TransATor, defined biosynthetic rules and generated biosynthetic models. P.M. designed the bioinformatics/chemoinformatics pipeline. P.M. and A.D. programmed TransATor. E.J.N.H. and A.D. validated TransATor and predicted compound structures. R.U. isolated compounds and performed structure elucidation. M.G., R.C. and G.C. provided genome sequences and bacterial strains. E.J.N.H. and J.P. wrote the manuscript with the help of all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–12, Supplementary Tables 1–10
Supplementary Note
Synthetic Procedures
Supplementary Dataset 1
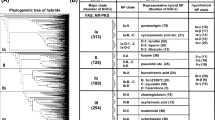
Maximum likelihood phylogenetic tree from a MUSCLE alignment of 655 KS sequences of all 54 characterized trans-AT PKS BGCs (as of December 2016).
Rights and permissions
About this article
Cite this article
Helfrich, E.J.N., Ueoka, R., Dolev, A. et al. Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat Chem Biol 15, 813–821 (2019). https://doi.org/10.1038/s41589-019-0313-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0313-7
This article is cited by
-
Global analysis of the biosynthetic chemical space of marine prokaryotes
Microbiome (2023)
-
Genome-based discovery and total synthesis of janustatins, potent cytotoxins from a plant-associated bacterium
Nature Chemistry (2022)
-
Strategies to access biosynthetic novelty in bacterial genomes for drug discovery
Nature Reviews Drug Discovery (2022)
-
An isotopic labeling approach linking natural products with biosynthetic gene clusters
Nature Chemical Biology (2022)
-
Automatic reconstruction of metabolic pathways from identified biosynthetic gene clusters
BMC Bioinformatics (2021)